Tissue tropism of swine influenza viruses and reassortants in ex vivo cultures of the human respiratory tract and conjunctiva
- PMID: 21880750
- PMCID: PMC3209323
- DOI: 10.1128/JVI.05662-11
Tissue tropism of swine influenza viruses and reassortants in ex vivo cultures of the human respiratory tract and conjunctiva
Abstract
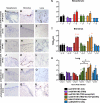
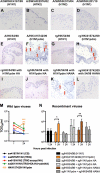
The 2009 pandemic influenza H1N1 (H1N1pdm) virus was generated by reassortment of swine influenza viruses of different lineages. This was the first influenza pandemic to emerge in over 4 decades and the first to occur after the realization that influenza pandemics arise from influenza viruses of animals. In order to understand the biological determinants of pandemic emergence, it is relevant to compare the tropism of different lineages of swine influenza viruses and reassortants derived from them with that of 2009 pandemic H1N1 (H1N1pdm) and seasonal influenza H1N1 viruses in ex vivo cultures of the human nasopharynx, bronchus, alveoli, and conjunctiva. We hypothesized that virus which can transmit efficiently between humans replicated well in the human upper airways. As previously reported, H1N1pdm and seasonal H1N1 viruses replicated efficiently in the nasopharyngeal, bronchial, and alveolar epithelium. In contrast, representative viruses from the classical swine (CS) (H1N1) lineage could not infect human respiratory epithelium; Eurasian avian-like swine (EA) (H1N1) viruses only infected alveolar epithelium and North American triple-reassortant (TRIG) viruses only infected the bronchial epithelium albeit inefficiently. Interestingly, a naturally occurring triple-reassortant swine virus, A/SW/HK/915/04 (H1N2), with a matrix gene segment of EA swine derivation (i.e., differing from H1N1pdm only in lacking a neuraminidase [NA] gene of EA derivation) readily infected and replicated in human nasopharyngeal and bronchial epithelia but not in the lung. A recombinant sw915 with the NA from H1N1pdm retained its tropism for the bronchus and acquired additional replication competence for alveolar epithelium. In contrast to H1N1pdm, none of the swine viruses tested nor seasonal H1N1 had tropism in human conjunctiva. Recombinant viruses generated by swapping the surface proteins (hemagglutinin and NA) of H1N1pdm and seasonal H1N1 virus demonstrated that these two gene segments together are key determinants of conjunctival tropism. Overall, these findings suggest that ex vivo cultures of the human respiratory tract provide a useful biological model for assessing the human health risk of swine influenza viruses.
Figures

References
-
- Dawood F. S., et al. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360: 2605–2615 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

