Characterization of Vibrio cholerae O1 El Tor biotype variant clinical isolates from Bangladesh and Haiti, including a molecular genetic analysis of virulence genes
- PMID: 21880975
- PMCID: PMC3209127
- DOI: 10.1128/JCM.01286-11
Characterization of Vibrio cholerae O1 El Tor biotype variant clinical isolates from Bangladesh and Haiti, including a molecular genetic analysis of virulence genes
Abstract
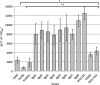
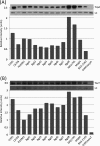
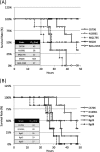
Vibrio cholerae serogroup O1, the causative agent of the diarrheal disease cholera, is divided into two biotypes: classical and El Tor. Both biotypes produce the major virulence factors toxin-coregulated pilus (TCP) and cholera toxin (CT). Although possessing genotypic and phenotypic differences, El Tor biotype strains displaying classical biotype traits have been reported and subsequently were dubbed El Tor variants. Of particular interest are reports of El Tor variants that produce various levels of CT, including levels typical of classical biotype strains. Here, we report the characterization of 10 clinical isolates from the International Centre for Diarrhoeal Disease Research, Bangladesh, and a representative strain from the 2010 Haiti cholera outbreak. We observed that all 11 strains produced increased CT (2- to 10-fold) compared to that of wild-type El Tor strains under in vitro inducing conditions, but they possessed various TcpA and ToxT expression profiles. Particularly, El Tor variant MQ1795, which produced the highest level of CT and very high levels of TcpA and ToxT, demonstrated hypervirulence compared to the virulence of El Tor wild-type strains in the infant mouse cholera model. Additional genotypic and phenotypic tests were conducted to characterize the variants, including an assessment of biotype-distinguishing characteristics. Notably, the sequencing of ctxB in some El Tor variants revealed two copies of classical ctxB, one per chromosome, contrary to previous reports that located ctxAB only on the large chromosome of El Tor biotype strains.
Figures

References
-
- Ansaruzzaman M., et al. 2007. Genetic diversity of El Tor strains of Vibrio cholerae O1 with hybrid traits isolated from Bangladesh and Mozambique. Int. J. Med. Microbiol. 297:443–449 - PubMed
-
- Barua D. 1992. History of cholera, p. 1–36 In Barua D., Greenough W. B., III (ed.), Cholera. Plenum Publishing Corporation, New York, NY
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

