Runx2 promotes both osteoblastogenesis and novel osteoclastogenic signals in ST2 mesenchymal progenitor cells
- PMID: 21881969
- PMCID: PMC5771409
- DOI: 10.1007/s00198-011-1728-5
Runx2 promotes both osteoblastogenesis and novel osteoclastogenic signals in ST2 mesenchymal progenitor cells
Abstract
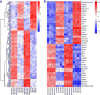
We profiled the global gene expression of a bone marrow-derived mesenchymal pluripotent cell line in response to Runx2 expression. Besides osteoblast differentiation, Runx2 promoted the osteoclastogenesis of co-cultured splenocytes. This was attributable to the upregulation of many novel osteoclastogenic genes and the downregulation of anti-osteoclastogenic genes.
Introduction: In addition to being a master regulator for osteoblast differentiation, Runx2 controls osteoblast-driven osteoclastogenesis. Previous studies profiling gene expression during osteoblast differentiation had limited focus on Runx2 or paid little attention to its role in mediating osteoblast-driven osteoclastogenesis.
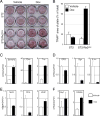
Methods: ST2/Rx2(dox), a bone marrow-derived mesenchymal pluripotent cell line that expresses Runx2 in response to Doxycycline (Dox), was used to profile Runx2-induced gene expression changes. Runx2-induced osteoblast differentiation was assessed based on alkaline phosphatase staining and expression of classical marker genes. Osteoclastogenic potential was evaluated by TRAP staining of osteoclasts that differentiated from primary murine splenocytes co-cultured with the ST2/Rx2(dox) cells. The BeadChip™ platform (Illumina) was used to interrogate genome-wide expression changes in ST2/Rx2(dox) cultures after treatment with Dox or vehicle for 24 or 48 h. Expression of selected genes was also measured by RT-qPCR.
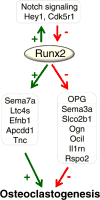
Results: Dox-mediated Runx2 induction in ST2 cells stimulated their own differentiation along the osteoblast lineage and the differentiation of co-cultured splenocytes into osteoclasts. The latter was attributable to the stimulation of osteoclastogenic genes such as Sema7a, Ltc4s, Efnb1, Apcdd1, and Tnc as well as the inhibition of anti-osteoclastogenic genes such as Tnfrsf11b (OPG), Sema3a, Slco2b1, Ogn, Clec2d (Ocil), Il1rn, and Rspo2.
Conclusion: Direct control of osteoblast differentiation and concomitant indirect control of osteoclast differentiation, both through the activity of Runx2 in pre-osteoblasts, constitute a novel mechanism of coordination with a potential crucial role in coupling bone formation and resorption.
Conflict of interest statement
Figures


References
-
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, Sato M, Okamoto R, Kitamura Y, Yoshiki S, Kishimoto T. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755–764. - PubMed
-
- Otto F, Thornell AP, Crompton T, Denzel A, Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen BR, Selby PB, Owen MJ. Cbfa1, a candidate gene for cleidocranial dysplasia syndrome, is essential for osteoblast differentiation and bone development. Cell. 1997;89:765–771. - PubMed
-
- Zhang YW, Yasui N, Kakazu N, Abe T, Takada K, Imai S, Sato M, Nomura S, Ochi T, Okuzumi S, Nogami H, Nagai T, Ohashi H, Ito Y. PEBP2alphaA/CBFA1 mutations in Japanese cleidocranial dysplasia patients. Gene. 2000;244:21–28. - PubMed
-
- Vaughan T, Pasco JA, Kotowicz MA, Nicholson GC, Morrison NA. Alleles of RUNX2/CBFA1 gene are associated with differences in bone mineral density and risk of fracture. J Bone Miner Res. 2002;17:1527–1534. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

