Long-lived states to monitor protein unfolding by proton NMR
- PMID: 21882334
- PMCID: PMC3368952
- DOI: 10.1002/cphc.201100365
Long-lived states to monitor protein unfolding by proton NMR
Abstract
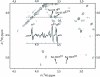
The relaxation of long-lived states (LLS) corresponds to the slow return to statistical thermal equilibrium between symmetric and antisymmetric proton spin states. This process is remarkably sensitive to the presence of external spins and can be used to obtain information about partial unfolding of proteins. We detected the appearance of a destabilized conformer of ubiquitin when urea is added to the protein in its native state. This conformer shows increased mobility in the C-terminus, which significantly extends the lifetimes of proton LLS magnetisation in Ser-65. These changes could not be detected by conventional measurements of T(1) and T(2) relaxation times of protons, and would hardly be sensed by carbon-13 or nitrogen-15 relaxation measurements. Conformers with similar dynamic and structural features, as revealed by LLS relaxation times, could be observed, in the absence of urea, in two ubiquitin mutants, L67S and L69S.
Copyright © 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures


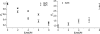
References
-
- Dobson CM, Karplus M. Curr Opin Struct Biol. 1999;9:92–101. - PubMed
-
- Fersht AR, Daggett V. Cell. 2002;108:573–582. - PubMed
-
- Ibarra-Molero B, Loladze VV, Makhatadze GI, Sanchez-Ruiz JM. Biochemistry (Mosc) 1999;38:8138–8149. - PubMed
-
- Pickart CM, Fushman D. Curr Opin Chem Biol. 2004;8:610–616. - PubMed
-
- Vijaykumar S, Bugg CE, Cook WJ. J Mol Biol. 1987;194:531–544. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

