Ubiquitination of substrates by esterification
- PMID: 21883762
- PMCID: PMC3973488
- DOI: 10.1111/j.1600-0854.2011.01269.x
Ubiquitination of substrates by esterification
Abstract
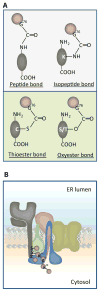
Post-translational modification by ubiquitination determines intracellular location and fate of numerous proteins, thus impacting a diverse array of physiologic functions. Past dogma has been that ubiquitin was only coupled to substrates by isopeptide bonds to internal lysine residues or less frequently peptide bonds to the N-terminus. Enigmatically, however, several proteins lacking lysines had been reported to retain ubiquitin-dependent fates. Resolution of this paradox was afforded by recent observations that ubiquitination of substrates can also occur on cysteine or serine and threonine residues by thio- or oxy-ester bond formation, respectively (collectively called esterification). Although chemically possible, these bonds were considered too labile to be of physiological relevance. In this review we discuss recent evidence for the ubiquitination of protein substrates by esterification and speculate on its mechanism and its physiological importance.
© 2011 John Wiley & Sons A/S.
Figures
References
-
- Ciechanover A, Ben Saadon R. N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol. 2004;14:103–106. - PubMed
-
- Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399434. - PubMed
-
- Coscoy L, Ganem D. PHD domains and E3 ubiquitin ligases: viruses make the connection. Trends Cell Biol. 2003;13:7–12. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

