Microbial rhodopsins on leaf surfaces of terrestrial plants
- PMID: 21883799
- PMCID: PMC3608849
- DOI: 10.1111/j.1462-2920.2011.02554.x
Microbial rhodopsins on leaf surfaces of terrestrial plants
Abstract
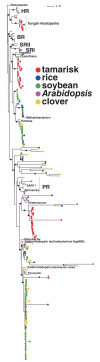
The above-ground surfaces of terrestrial plants, the phyllosphere, comprise the main interface between the terrestrial biosphere and solar radiation. It is estimated to host up to 10(26) microbial cells that may intercept part of the photon flux impinging on the leaves. Based on 454-pyrosequencing-generated metagenome data, we report on the existence of diverse microbial rhodopsins in five distinct phyllospheres from tamarisk (Tamarix nilotica), soybean (Glycine max), Arabidopsis (Arabidopsis thaliana), clover (Trifolium repens) and rice (Oryza sativa). Our findings, for the first time describing microbial rhodopsins from non-aquatic habitats, point towards the potential coexistence of microbial rhodopsin-based phototrophy and plant chlorophyll-based photosynthesis, with the different pigments absorbing non-overlapping fractions of the light spectrum.
© 2011 Society for Applied Microbiology and Blackwell Publishing Ltd.
Figures



References
-
- Atamna-Ismaeel N, Sabehi G, Sharon I, Witzel K-P, Labrenz M, Jürgens K, et al. Widespread distribution of proteorhodopsins in freshwater and brackish ecosystems. ISME J. 2008;2:656–662. - PubMed
-
- Ballaré CL, Scopel AL, Sánchez RA. Far-red radiation reflected from adjacent leaves: an early signal of competition in plant canopies. Science. 1990;247:329–332. - PubMed
-
- Beattie GA, Lindow SE. Bacterial colonization of leaves: a spectrum of strategies. Phytopathol. 1999;89:353–359. - PubMed
-
- Béjà O, Aravind L, Koonin EV, Suzuki MT, Hadd A, Nguyen LP, et al. Bacterial rhodopsin: evidence for a new type of phototrophy in the sea. Science. 2000;289:1902–1906. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

