Factors that influence the response of the LysR type transcriptional regulators to aromatic compounds
- PMID: 21884597
- PMCID: PMC3180648
- DOI: 10.1186/1471-2091-12-49
Factors that influence the response of the LysR type transcriptional regulators to aromatic compounds
Abstract
Background: The transcriptional regulators DntR, NagR and NtdR have a high sequence identity and belong to the large family of LysR type transcriptional regulators (LTTRs). These three regulators are all involved in regulation of genes identified in pathways for degradation of aromatic compounds. They activate the transcription of these genes in the presence of an inducer, but the inducer specificity profiles are different.
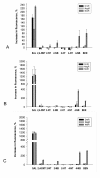
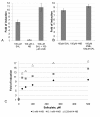
Results: The results from this study show that NtdR has the broadest inducer specificity, responding to several nitro-aromatic compounds. Mutational studies of residues that differ between DntR, NagR and NtdR suggest that a number of specific residues are involved in the broader inducer specificity of NtdR when compared to DntR and NagR. The inducer response was also investigated as a function of the experimental conditions and a number of parameters such as the growth media, plasmid arrangement of the LTTR-encoding genes, promoter and gfp reporter gene, and the presence of a His6-tag were shown to affect the inducer response in E.coli DH5α. Furthermore, the response upon addition of both salicylate and 4-nitrobenzoate to the growth media was larger than the sum of responses upon addition of each of the compounds, which suggests the presence of a secondary binding site, as previously reported for other LTTRs.
Conclusions: Optimization of the growth conditions and gene arrangement resulted in improved responses to nitro-aromatic inducers. The data also suggests the presence of a previously unknown secondary binding site in DntR, analogous to that of BenM.
Figures



Similar articles
-
Reconstructing the evolutionary history of nitrotoluene detection in the transcriptional regulator NtdR.Mol Microbiol. 2009 Nov;74(4):826-43. doi: 10.1111/j.1365-2958.2009.06904.x. Epub 2009 Oct 22. Mol Microbiol. 2009. PMID: 19849778 Free PMC article.
-
Crystal structures of DntR inducer binding domains in complex with salicylate offer insights into the activation of LysR-type transcriptional regulators.Mol Microbiol. 2011 Jul;81(2):354-67. doi: 10.1111/j.1365-2958.2011.07673.x. Epub 2011 Jun 22. Mol Microbiol. 2011. PMID: 21692874
-
Crystal structure of the DNA-binding domain of the LysR-type transcriptional regulator CbnR in complex with a DNA fragment of the recognition-binding site in the promoter region.FEBS J. 2018 Mar;285(5):977-989. doi: 10.1111/febs.14380. Epub 2018 Jan 28. FEBS J. 2018. PMID: 29323785
-
Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins.Microbiology (Reading). 2008 Dec;154(Pt 12):3609-3623. doi: 10.1099/mic.0.2008/022772-0. Microbiology (Reading). 2008. PMID: 19047729 Review.
-
Molecular biology of the LysR family of transcriptional regulators.Annu Rev Microbiol. 1993;47:597-626. doi: 10.1146/annurev.mi.47.100193.003121. Annu Rev Microbiol. 1993. PMID: 8257110 Review.
Cited by
-
Regulation of the aceI multidrug efflux pump gene in Acinetobacter baumannii.J Antimicrob Chemother. 2018 Jun 1;73(6):1492-1500. doi: 10.1093/jac/dky034. J Antimicrob Chemother. 2018. PMID: 29481596 Free PMC article.
-
Use of Substrate-Induced Gene Expression in Metagenomic Analysis of an Aromatic Hydrocarbon-Contaminated Soil.Appl Environ Microbiol. 2015 Nov 20;82(3):897-909. doi: 10.1128/AEM.03306-15. Print 2016 Feb 1. Appl Environ Microbiol. 2015. PMID: 26590287 Free PMC article.
-
The solution configurations of inactive and activated DntR have implications for the sliding dimer mechanism of LysR transcription factors.Sci Rep. 2016 Jan 28;6:19988. doi: 10.1038/srep19988. Sci Rep. 2016. PMID: 26817994 Free PMC article.
-
A novel LysR-type regulator negatively affects biosynthesis of the immunosuppressant brasilicardin.Eng Life Sci. 2020 Nov 4;21(1-2):4-18. doi: 10.1002/elsc.202000038. eCollection 2021 Jan. Eng Life Sci. 2020. PMID: 33531886 Free PMC article.
-
Optochemical Control of Bacterial Gene Expression: Novel Photocaged Compounds for Different Promoter Systems.Chembiochem. 2022 Jan 5;23(1):e202100467. doi: 10.1002/cbic.202100467. Epub 2021 Dec 2. Chembiochem. 2022. PMID: 34750949 Free PMC article.
References
-
- Maddocks SE, Oyston PC. Structure and function of the LysR-type transcriptional regulator (LTTR) family proteins. Microbiology. 2008;154(Pt 12):3609–3623. - PubMed
-
- Sainsbury S, Lane LA, Ren J, Gilbert RJ, Saunders NJ, Robinson CV, Stuart DI, Owens RJ. The structure of CrgA from Neisseria meningitidis reveals a new octameric assembly state for LysR transcriptional regulators. Nucleic Acids Res. 2009;37(14):4545–4558. doi: 10.1093/nar/gkp445. - DOI - PMC - PubMed
-
- Zhou X, Lou Z, Fu S, Yang A, Shen H, Li Z, Feng Y, Bartlam M, Wang H, Rao Z. Crystal structure of ArgP from Mycobacterium tuberculosis confirms two distinct conformations of full-length LysR transcriptional regulators and reveals its function in DNA binding and transcriptional regulation. J Mol Biol. 2010;396(4):1012–1024. doi: 10.1016/j.jmb.2009.12.033. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

