GO-based functional dissimilarity of gene sets
- PMID: 21884611
- PMCID: PMC3248071
- DOI: 10.1186/1471-2105-12-360
GO-based functional dissimilarity of gene sets
Abstract
Background: The Gene Ontology (GO) provides a controlled vocabulary for describing the functions of genes and can be used to evaluate the functional coherence of gene sets. Many functional coherence measures consider each pair of gene functions in a set and produce an output based on all pairwise distances. A single gene can encode multiple proteins that may differ in function. For each functionality, other proteins that exhibit the same activity may also participate. Therefore, an identification of the most common function for all of the genes involved in a biological process is important in evaluating the functional similarity of groups of genes and a quantification of functional coherence can helps to clarify the role of a group of genes working together.
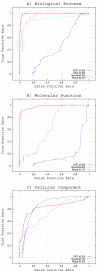
Results: To implement this approach to functional assessment, we present GFD (GO-based Functional Dissimilarity), a novel dissimilarity measure for evaluating groups of genes based on the most relevant functions of the whole set. The measure assigns a numerical value to the gene set for each of the three GO sub-ontologies.
Conclusions: Results show that GFD performs robustly when applied to gene set of known functionality (extracted from KEGG). It performs particularly well on randomly generated gene sets. An ROC analysis reveals that the performance of GFD in evaluating the functional dissimilarity of gene sets is very satisfactory. A comparative analysis against other functional measures, such as GS2 and those presented by Resnik and Wang, also demonstrates the robustness of GFD.
Figures

References
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature Genetics. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
-
- Pontius J, Wagner L, Schuler G. UniGene: a unified view of the transcriptome. The NCBI Handbook. Bethesda (MD): National Center for Biotechnology Information. 2003.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

