Expression profiling reveals differential gene induction underlying specific and non-specific memory for pheromones in mice
- PMID: 21884744
- PMCID: PMC3206202
- DOI: 10.1016/j.neuint.2011.08.009
Expression profiling reveals differential gene induction underlying specific and non-specific memory for pheromones in mice
Abstract
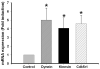
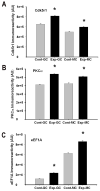
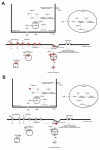
Memory for the mating male's pheromones in female mice is thought to require synaptic changes in the accessory olfactory bulb (AOB). Induction of this memory depends on release of glutamate in response to pheromonal exposure coincident with release of norepinephrine (NE) in the AOB following mating. A similar memory for pheromones can also be induced artificially by local infusion of the GABA(A) receptor antagonist bicuculline into the AOB. The natural memory formed by exposure to pheromones during mating is specific to the pheromones sensed by the female during mating. In contrast, the artificial memory induced by bicuculline is non-specific and results in the female mice recognizing all pheromones as if they were from the mating male. Although protein synthesis has been shown to be essential for development of pheromone memory, the gene expression cascades critical for memory formation are not known. We investigated changes in gene expression in the AOB using oligonucleotide microarrays during mating-induced pheromone memory (MIPM) as well as bicuculline-induced pheromone memory (BIPM). We found the set of genes induced during MIPM and BIPM are largely non-overlapping and Ingenuity Pathway Analysis revealed that the signaling pathways in MIPM and BIPM also differ. The products of genes induced during MIPM are associated with synaptic function, indicating the possibility of modification at specific synapses, while those induced during BIPM appear to possess neuron-wide functions, which would be consistent with global cellular changes. Thus, these results begin to provide a mechanistic explanation for specific and non-specific memories induced by pheromones and bicuculline infusion respectively.
Copyright © 2011 Elsevier B.V. All rights reserved.
Figures




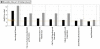


References
-
- Bedford FK, Kittler JT, Muller E, Thomas P, Uren JM, Merlo D, Wisden W, Triller A, Smart TG, Moss SJ. GABA(A) receptor cell surface number and subunit stability are regulated by the ubiquitin-like protein Plic-1. Nat. Neurosci. 2001;4:908–916. - PubMed
-
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society. Series B (Methodological) 1995;57:289–300.
-
- Bramham CR, Messaoudi E. BDNF function in adult synaptic plasticity: the synaptic consolidation hypothesis. Prog. Neurobiol. 2005;76:99–125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

