TET family proteins and their role in stem cell differentiation and transformation
- PMID: 21885017
- PMCID: PMC3244690
- DOI: 10.1016/j.stem.2011.08.007
TET family proteins and their role in stem cell differentiation and transformation
Abstract
One of the main regulators of gene expression during embryogenesis and stem cell differentiation is DNA methylation. The recent identification of hydroxymethylcytosine (5hmC) as a novel epigenetic mark sparked an intense effort to characterize its specialized enzymatic machinery and to understand the biological significance of 5hmC. The recent discovery of recurrent deletions and somatic mutations in the TET gene family, which includes proteins that can hydroxylate methylcytosine (5mC), in a large fraction of myeloid malignancies further suggested a key role for dynamic DNA methylation changes in the regulation of stem cell differentiation and transformation.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


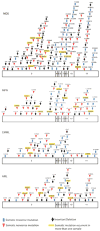
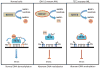
References
-
- Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, et al. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. - PubMed
-
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U54 CA143798/CA/NCI NIH HHS/United States
- R01 CA133379/CA/NCI NIH HHS/United States
- R01CA133379/CA/NCI NIH HHS/United States
- R21CA141399/CA/NCI NIH HHS/United States
- R01 CA149655/CA/NCI NIH HHS/United States
- R01 CA105129/CA/NCI NIH HHS/United States
- HHMI/Howard Hughes Medical Institute/United States
- R21 CA141399/CA/NCI NIH HHS/United States
- R01CA149655/CA/NCI NIH HHS/United States
- R01CA105129/CA/NCI NIH HHS/United States
- 1R01CA138234-01/CA/NCI NIH HHS/United States
- R01GM088847/GM/NIGMS NIH HHS/United States
- R01 CA138234/CA/NCI NIH HHS/United States
- U54CA143798-01/CA/NCI NIH HHS/United States
- R01 GM088847/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

