Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation
- PMID: 21885282
- PMCID: PMC3175335
- DOI: 10.1016/j.cub.2011.08.005
Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation
Abstract
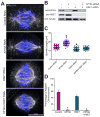
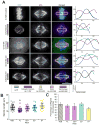
The mitotic spindle is a macromolecular structure utilized to properly align and segregate sister chromatids to two daughter cells. During mitosis, the spindle maintains a constant length, even though the spindle microtubules (MTs) are constantly undergoing polymerization and depolymerization [1]. Members of the kinesin-8 family are important for the regulation of spindle length and for chromosome positioning [2-9]. Kinesin-8 proteins are length-specific, plus-end-directed motors that are proposed to be either MT depolymerases [3, 4, 8, 10, 11] or MT capping proteins [12]. How Kif18A uses its destabilization activity to control spindle morphology is not known. We found that Kif18A controls spindle length independently of its role in chromosome positioning. The ability of Kif18A to control spindle length is mediated by an ATP-independent MT binding site at the C-terminal end of the Kif18A tail that has a strong affinity for MTs in vitro and in cells. We used computational modeling to ask how modulating the motility or binding properties of Kif18A would affect its activity. Our modeling predicts that both fast motility and a low off rate from the MT end are important for Kif18A function. In addition, our studies provide new insight into how depolymerizing and capping enzymes can lead to MT destabilization.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures


References
-
- Goshima G, Scholey JM. Control of mitotic spindle length. Annu Rev Cell Dev Biol. 2010;26:21–57. - PubMed
-
- Goshima G, Wollman R, Stuurman N, Scholey JM, Vale RD. Length control of the metaphase spindle. Curr Biol. 2005;15:1979–1988. - PubMed
-
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. - PubMed
-
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. - PubMed
-
- Savoian MS, Gatt MK, Riparbelli MG, Callaini G, Glover DM. Drosophila Klp67A is required for proper chromosome congression and segregation during meiosis I. J Cell Sci. 2004;117:3669–3677. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

