Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network
- PMID: 21886106
- PMCID: PMC3654671
- DOI: 10.1038/nprot.2011.372
Systematic prediction of gene function in Arabidopsis thaliana using a probabilistic functional gene network
Abstract
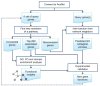
AraNet is a functional gene network for the reference plant Arabidopsis and has been constructed in order to identify new genes associated with plant traits. It is highly predictive for diverse biological pathways and can be used to prioritize genes for functional screens. Moreover, AraNet provides a web-based tool with which plant biologists can efficiently discover novel functions of Arabidopsis genes (http://www.functionalnet.org/aranet/). This protocol explains how to conduct network-based prediction of gene functions using AraNet and how to interpret the prediction results. Functional discovery in plant biology is facilitated by combining candidate prioritization by AraNet with focused experimental tests.
Figures







References
-
- Lehner B, Lee I. Network-guided genetic screening: building, testing and using gene networks to predict gene function. Brief Funct. Genomic Proteomic. 2008;7:217–227. - PubMed
-
- Alonso JM, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. - PubMed
-
- Lee I, Date SV, Adai AT, Marcotte EM. A probabilistic functional network of yeast genes. Science. 2004;306:1555–1558. - PubMed
-
- Marcotte EM, Pellegrini M, Thompson MJ, Yeates TO, Eisenberg D. A combined algorithm for genome-wide prediction of protein function. Nature. 1999;402:83–86. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

