One carbon metabolism in SAR11 pelagic marine bacteria
- PMID: 21886845
- PMCID: PMC3160333
- DOI: 10.1371/journal.pone.0023973
One carbon metabolism in SAR11 pelagic marine bacteria
Abstract
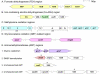
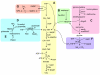
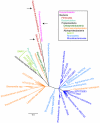
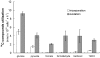
The SAR11 Alphaproteobacteria are the most abundant heterotrophs in the oceans and are believed to play a major role in mineralizing marine dissolved organic carbon. Their genomes are among the smallest known for free-living heterotrophic cells, raising questions about how they successfully utilize complex organic matter with a limited metabolic repertoire. Here we show that conserved genes in SAR11 subgroup Ia (Candidatus Pelagibacter ubique) genomes encode pathways for the oxidation of a variety of one-carbon compounds and methyl functional groups from methylated compounds. These pathways were predicted to produce energy by tetrahydrofolate (THF)-mediated oxidation, but not to support the net assimilation of biomass from C1 compounds. Measurements of cellular ATP content and the oxidation of (14)C-labeled compounds to (14)CO(2) indicated that methanol, formaldehyde, methylamine, and methyl groups from glycine betaine (GBT), trimethylamine (TMA), trimethylamine N-oxide (TMAO), and dimethylsulfoniopropionate (DMSP) were oxidized by axenic cultures of the SAR11 strain Ca. P. ubique HTCC1062. Analyses of metagenomic data showed that genes for C1 metabolism occur at a high frequency in natural SAR11 populations. In short term incubations, natural communities of Sargasso Sea microbial plankton expressed a potential for the oxidation of (14)C-labeled formate, formaldehyde, methanol and TMAO that was similar to cultured SAR11 cells and, like cultured SAR11 cells, incorporated a much larger percentage of pyruvate and glucose (27-35%) than of C1 compounds (2-6%) into biomass. Collectively, these genomic, cellular and environmental data show a surprising capacity for demethylation and C1 oxidation in SAR11 cultures and in natural microbial communities dominated by SAR11, and support the conclusion that C1 oxidation might be a significant conduit by which dissolved organic carbon is recycled to CO(2) in the upper ocean.
Conflict of interest statement
Figures


References
-
- Henderson JF. Teaching one-carbon metabolism. Biochem Educ. 1979;7:51–52.
-
- McDowell LR. Vitamins in animal and human nutrition: Iowa State University Press. 2000. 487
-
- Chistoserdova L, Vorholt JA, Thauer RK, Lidstrom ME. C1 transfer enzymes and coenzymes linking methylotrophic bacteria and methanogenic Archaea. Science. 1998;281:99–102. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

