Screening of an E. coli O157:H7 Bacterial Artificial Chromosome Library by Comparative Genomic Hybridization to Identify Genomic Regions Contributing to Growth in Bovine Gastrointestinal Mucus and Epithelial Cell Colonization
- PMID: 21887152
- PMCID: PMC3157008
- DOI: 10.3389/fmicb.2011.00168
Screening of an E. coli O157:H7 Bacterial Artificial Chromosome Library by Comparative Genomic Hybridization to Identify Genomic Regions Contributing to Growth in Bovine Gastrointestinal Mucus and Epithelial Cell Colonization
Abstract
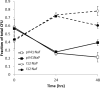
Enterohemorrhagic E. coli (EHEC) O157:H7 can cause serious gastrointestinal and systemic disease in humans following direct or indirect exposure to ruminant feces containing the bacterium. The main colonization site of EHEC O157:H7 in cattle is the terminal rectum where the bacteria intimately attach to the epithelium and multiply in the intestinal mucus. This study aimed to identify genomic regions of EHEC O157:H7 that contribute to colonization and multiplication at this site. A bacterial artificial chromosome (BAC) library was generated from a derivative of the sequenced E. coli O157:H7 Sakai strain. The library contains 1152 clones averaging 150 kbp. To verify the library, clones containing a complete locus of enterocyte effacement (LEE) were identified by DNA hybridization. In line with a previous report, these did not confer a type III secretion (T3S) capacity to the K-12 host strain. However, conjugation of one of the BAC clones into a strain containing a partial LEE deletion restored T3S. Three hundred eighty-four clones from the library were subjected to two different selective screens; one involved three rounds of adherence assays to bovine primary rectal epithelial cells while the other competed the clones over three rounds of growth in bovine rectal mucus. The input strain DNA was then compared with the selected strains using comparative genomic hybridization (CGH) on an E. coli microarray. The adherence assay enriched for pO157 DNA indicating the importance of this plasmid for colonization of rectal epithelial cells. The mucus assay enriched for multiple regions involved in carbohydrate utilization, including hexuronate uptake, indicating that these regions provide a competitive growth advantage in bovine mucus. This BAC-CGH approach provides a positive selection screen that complements negative selection transposon-based screens. As demonstrated, this may be of particular use for identifying genes with redundant functions such as adhesion and carbon metabolism.
Keywords: EHEC; bacterial artificial chromosome; comparative genomic hybridization; locus of enterocyte effacement; mucus; sugar utilization.
Figures
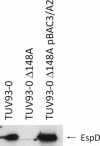

References
-
- Chang D. E., Smalley D. J., Tucker D. L., Leatham M. P., Norris W. E., Stevenson S. J., Anderson A. B., Grissom J. E., Laux D. C., Cohen P. S., Conway T. (2004). Carbon nutrition of Escherichia coli in the mouse intestine. Proc. Natl. Acad. Sci. U.S.A. 101, 7427–7432 10.1073/pnas.0408439102 - DOI - PMC - PubMed
-
- Conway T., Krogfelt K. A., Cohen P. S. (2004). The Life of Commensal Escherichia coli in the Mammalian Intestine. Washington, DC: ASM Press - PubMed
-
- Conway T., Krogfelt K. A., Cohen P. S. (2007). “Escherichia coli at the intestinal mucosal surface,” in Virulence Mechanisms of Bacterial Pathogens, 4th Edn, eds Brogden K. A., Minion F. C., Cornick N., Stanton T. B., Zhang Q., Nolan L. K., Wannemuehler M. J. (Washington, DC: ASM Press; ), 175–196
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

