The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia
- PMID: 21890456
- PMCID: PMC3207001
- DOI: 10.1158/1078-0432.CCR-11-0785
The prognostic significance of various 13q14 deletions in chronic lymphocytic leukemia
Abstract
Purpose: To further our understanding of the biology and prognostic significance of various chromosomal 13q14 deletions in chronic lymphocytic leukemia (CLL).
Experimental design: We analyzed data from SNP 6.0 arrays to define the anatomy of various 13q14 deletions in a cohort of 255 CLL patients and have correlated two subsets of 13q14 deletions (type I exclusive of RB1 and type II inclusive of RB1) with patient survival. Furthermore, we measured the expression of the 13q14-resident microRNAs by quantitative PCR (Q-PCR) in 242 CLL patients and subsequently assessed their prognostic significance. We sequenced all coding exons of RB1 in patients with monoallelic RB1 deletion and have sequenced the 13q14-resident miR locus in all patients.
Results: Large 13q14 (type II) deletions were detected in approximately 20% of all CLL patients and were associated with shortened survival. A strong association between 13q14 type II deletions and elevated genomic complexity, as measured through CLL-FISH or SNP 6.0 array profiling, was identified, suggesting that these lesions may contribute to CLL disease evolution through genomic destabilization. Sequence and copy number analysis of the RB1 gene identified a small CLL subset that is RB1 null. Finally, neither the expression levels of the 13q14-resident microRNAs nor the degree of 13q14 deletion, as measured through SNP 6.0 array-based copy number analysis, had significant prognostic importance.
Conclusions: Our data suggest that the clinical course of CLL is accelerated in patients with large (type II) 13q14 deletions that span the RB1 gene, therefore justifying routine identification of 13q14 subtypes in CLL management.
©2011 AACR
Figures

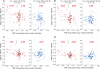



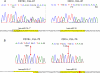
References
-
- Dohner H, Stilgenbauer S, Benner A, Leupolt E, Krober A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343:1910–6. - PubMed
-
- Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352:804–15. - PubMed
-
- Shanafelt TD, Hanson C, Dewald GW, Witzig TE, LaPlant B, Abrahamzon J, et al. Karyotype evolution on fluorescent in situ hybridization analysis is associated with short survival in patients with chronic lymphocytic leukemia and is related to CD49d expression. J Clin Oncol. 2008;26:e5–6. - PMC - PubMed
-
- Juliusson G, Oscier DG, Fitchett M, Ross FM, Stockdill G, Mackie MJ, et al. Prognostic subgroups in B-cell chronic lymphocytic leukemia defined by specific chromosomal abnormalities. N Engl J Med. 1990;323:720–4. - PubMed
-
- Peterson LC, Lindquist LL, Church S, Kay NE. Frequent clonal abnormalities of chromosome band 13q14 in B-cell chronic lymphocytic leukemia: multiple clones, subclones, and nonclonal alterations in 82 midwestern patients. Genes Chromosomes Cancer. 1992;4:273–80. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

