Divergence of structure and function in the haloacid dehalogenase enzyme superfamily: Bacteroides thetaiotaomicron BT2127 is an inorganic pyrophosphatase
- PMID: 21894910
- PMCID: PMC3342813
- DOI: 10.1021/bi201181q
Divergence of structure and function in the haloacid dehalogenase enzyme superfamily: Bacteroides thetaiotaomicron BT2127 is an inorganic pyrophosphatase
Abstract
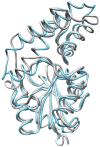
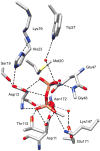
The explosion of protein sequence information requires that current strategies for function assignment evolve to complement experimental approaches with computationally based function prediction. This necessitates the development of strategies based on the identification of sequence markers in the form of specificity determinants and a more informed definition of orthologues. Herein, we have undertaken the function assignment of the unknown haloalkanoate dehalogenase superfamily member BT2127 (Uniprot accession code Q8A5 V9) from Bacteroides thetaiotaomicron using an integrated bioinformatics-structure-mechanism approach. The substrate specificity profile and steady-state rate constants of BT2127 (with a k(cat)/K(m) value for pyrophosphate of ~1 × 10(5) M(-1) s(-1)), together with the gene context, support the assigned in vivo function as an inorganic pyrophosphatase. The X-ray structural analysis of wild-type BT2127 and several variants generated by site-directed mutagenesis shows that substrate discrimination is based, in part, on active site space restrictions imposed by the cap domain (specifically by residues Tyr76 and Glu47). Structure-guided site-directed mutagenesis coupled with kinetic analysis of the mutant enzymes identified the residues required for catalysis, substrate binding, and domain-domain association. On the basis of this structure-function analysis, the catalytic residues Asp11, Asp13, Thr113, and Lys147 as well the metal binding residues Asp171, Asn172, and Glu47 were used as markers to confirm BT2127 orthologues identified via sequence searches. This bioinformatic analysis demonstrated that the biological range of BT2127 orthologue is restricted to the phylum Bacteroidetes/Chlorobi. The key structural determinants in the divergence of BT2127 and its closest homologue, β-phosphoglucomutase, control the leaving group size (phosphate vs glucose phosphate) and the position of the Asp acid/base in the open versus closed conformations. HADSF pyrophosphatases represent a third mechanistic and fold type for bacterial pyrophosphatases.
© 2011 American Chemical Society
Figures



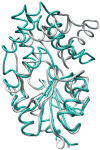

Similar articles
-
Insights into the functional divergence of the haloacid dehalogenase superfamily from phosphomonoesterase to inorganic pyrophosphatase.Arch Biochem Biophys. 2021 Jul 15;705:108896. doi: 10.1016/j.abb.2021.108896. Epub 2021 Apr 30. Arch Biochem Biophys. 2021. PMID: 33940035
-
The His23 and Lys79 pair determines the high catalytic efficiency of the inorganic pyrophosphatase of the haloacid dehalogenase superfamily.Biochim Biophys Acta Gen Subj. 2022 Jun;1866(6):130128. doi: 10.1016/j.bbagen.2022.130128. Epub 2022 Mar 10. Biochim Biophys Acta Gen Subj. 2022. PMID: 35278619
-
The X-ray crystallographic structure and specificity profile of HAD superfamily phosphohydrolase BT1666: comparison of paralogous functions in B. thetaiotaomicron.Proteins. 2011 Nov;79(11):3099-107. doi: 10.1002/prot.23137. Epub 2011 Aug 30. Proteins. 2011. PMID: 21989931 Free PMC article.
-
Active site interactions in oligomeric structures of inorganic pyrophosphatases.Biochemistry (Mosc). 2000 Mar;65(3):361-72. Biochemistry (Mosc). 2000. PMID: 10739480 Review.
-
Topological variation in the evolution of new reactions in functionally diverse enzyme superfamilies.Curr Opin Struct Biol. 2011 Jun;21(3):391-7. doi: 10.1016/j.sbi.2011.03.007. Epub 2011 Apr 1. Curr Opin Struct Biol. 2011. PMID: 21458983 Free PMC article. Review.
Cited by
-
Pyrophosphate-fueled Na+ and H+ transport in prokaryotes.Microbiol Mol Biol Rev. 2013 Jun;77(2):267-76. doi: 10.1128/MMBR.00003-13. Microbiol Mol Biol Rev. 2013. PMID: 23699258 Free PMC article. Review.
-
Roles of type II H+-PPases and PPsPase1/PECP2 in early developmental stages and PPi homeostasis of Arabidopsis thaliana.Front Plant Sci. 2023 Jan 27;14:1031426. doi: 10.3389/fpls.2023.1031426. eCollection 2023. Front Plant Sci. 2023. PMID: 36778688 Free PMC article.
-
Panoramic view of a superfamily of phosphatases through substrate profiling.Proc Natl Acad Sci U S A. 2015 Apr 21;112(16):E1974-83. doi: 10.1073/pnas.1423570112. Epub 2015 Apr 6. Proc Natl Acad Sci U S A. 2015. PMID: 25848029 Free PMC article.
-
Cystathionine β-synthase (CBS) domains confer multiple forms of Mg2+-dependent cooperativity to family II pyrophosphatases.J Biol Chem. 2014 Aug 15;289(33):22865-22876. doi: 10.1074/jbc.M114.589473. Epub 2014 Jul 1. J Biol Chem. 2014. PMID: 24986864 Free PMC article.
-
A toxin-antitoxin system provides phage defense via DNA damage and repair.Nat Commun. 2025 Apr 1;16(1):3141. doi: 10.1038/s41467-025-58540-9. Nat Commun. 2025. PMID: 40169592 Free PMC article.
References
-
- Allen KN, Dunaway-Mariano D. Phosphoryl group transfer: evolution of a catalytic scaffold. Trends Biochem Sci. 2004;29:495–503. - PubMed
-
- Burroughs AM, Allen KN, Dunaway-Mariano D, Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J Mol Biol. 2006;361:1003–1034. - PubMed
-
- Kuznetsova E, Proudfoot M, Gonzalez CF, Brown G, Omelchenko MV, Borozan I, Carmel L, Wolf YI, Mori H, Savchenko AV, Arrowsmith CH, Koonin EV, Edwards AM, Yakunin AF. Genome-wide analysis of substrate specificities of the Escherichia coli haloacid dehalogenase-like phosphatase family. J Biol Chem. 2006;281:36149–36161. - PubMed
-
- Wang W, Cho HS, Kim R, Jancarik J, Yokota H, Nguyen HH, Grigoriev IV, Wemmer DE, Kim SH. Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic “snapshots” of intermediate states. J Mol Biol. 2002;319:421–431. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

