Effects of palmitoylation of Cys(415) in helix 8 of the CB(1) cannabinoid receptor on membrane localization and signalling
- PMID: 21895628
- PMCID: PMC3423250
- DOI: 10.1111/j.1476-5381.2011.01658.x
Effects of palmitoylation of Cys(415) in helix 8 of the CB(1) cannabinoid receptor on membrane localization and signalling
Abstract
Background and purpose: The CB(1) cannabinoid receptor is regulated by its association with membrane microdomains such as lipid rafts. Here, we investigated the role of palmitoylation of the CB(1) receptor by analysing the functional consequences of site-specific mutation of Cys(415) , the likely site of palmitoylation at the end of helix 8, in terms of membrane association, raft targeting and signalling.
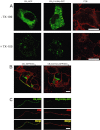
Experimental approach: The palmitoylation state of CB(1) receptors in rat forebrain was assessed by depalmitoylation/repalmitoylation experiments. Cys(415) was replaced with alanine by site-directed mutagenesis. Green fluorescence protein chimeras of both wild-type and mutant receptors were transiently expressed and functionally characterized in SH-SY5Y cells and HEK-293 cells by means of confocal microscopy, cytofluorimetry and competitive binding assays. Confocal fluorescence recovery after photobleaching was used to assess receptor membrane dynamics, whereas signalling activity was assessed by [(35) S]GTPγS, cAMP and co-immunoprecipitation assays.
Key results: Endogenous CB(1) receptors in rat brain were palmitoylated. Mutation of Cys(415) prevented the palmitoylation of the receptor in transfected cells and reduced its recruitment to plasma membrane and lipid rafts; it also increased protein diffusional mobility. The same mutation markedly reduced the functional coupling of CB(1) receptors with G-proteins and adenylyl cyclase, whereas depalmitoylation abolished receptor association with a specific subset of G-proteins.
Conclusions and implications: CB(1) receptors were post-translationally modified by palmitoylation. Mutation of Cys(415) provides a receptor that is functionally impaired in terms of membrane targeting and signalling.
Linked articles: This article is part of a themed section on Cannabinoids in Biology and Medicine. To view the other articles in this section visit http://dx.doi.org/10.1111/bph.2012.165.issue-8. To view Part I of Cannabinoids in Biology and Medicine visit http://dx.doi.org/10.1111/bph.2011.163.issue-7.
© 2011 The Authors. British Journal of Pharmacology © 2011 The British Pharmacological Society.
Figures

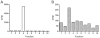




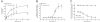
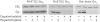
References
-
- Anderson RG, Jacobson K. A role for lipid shells in targetinG-proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–1825. - PubMed
-
- Bari M, Battista N, Fezza F, Finazzi-Agrò A, Maccarrone M. Lipid rafts control signaling of type-1 cannabinoid receptors in neuronal cells. Implications for anandamide-induced apoptosis. J Biol Chem. 2005;280:12212–12220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

