Histone octamer trans-transfer: a signature mechanism of ATP-dependent chromatin remodelling unravelled in wheat nuclear extract
- PMID: 21896571
- PMCID: PMC3197459
- DOI: 10.1093/aob/mcr232
Histone octamer trans-transfer: a signature mechanism of ATP-dependent chromatin remodelling unravelled in wheat nuclear extract
Abstract
Background and scope: In eukaryotes, chromatin remodelling complexes are shown to be responsible for nucleosome mobility, leading to increased accessibility of DNA for DNA binding proteins. Although the existence of such complexes in plants has been surmised mainly at the genetic level from bioinformatics studies and analysis of mutants, the biochemical existence of such complexes has remained unexplored.
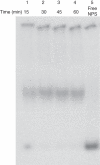
Methods: Histone H1-depleted donor chromatin was prepared by micrococcal nuclease digestion of wheat nuclei and fractionation by exclusion chromatography. Nuclear extract was partially purified by cellulose phosphate ion exchange chromatography. Histone octamer trans-transfer activity was analysed using the synthetic nucleosome positioning sequence in the absence and presence of ATP and its analogues. ATPase activity was measured as (32)Pi released using liquid scintillation counting.
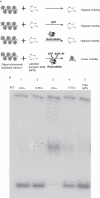
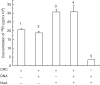
Key results: ATP-dependent histone octamer trans-transfer activity, partially purified from wheat nuclei using cellulose phosphate, showed ATP-dependent octamer displacement in trans from the H1-depleted native donor chromatin of wheat to the labelled synthetic nucleosome positioning sequence. It also showed nucleosome-dependent ATPase activity. Substitution of ATP by ATP analogues, namely ATPγS, AMP-PNP and ADP abolished the octamer trans-transfer, indicating the requirement of ATP hydrolysis for this activity.
Conclusions: ATP-dependent histone octamer transfer in trans is a recognized activity of chromatin remodelling complexes required for chromatin structure dynamics in non-plant species. Our results suggested that wheat nuclei also possess a typical chromatin remodelling activity, similar to that in other eukaryotes. This is the first report on chromatin remodelling activity in vitro from plants.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

