Transcriptional regulation of xenobiotic detoxification in Drosophila
- PMID: 21896655
- PMCID: PMC3175716
- DOI: 10.1101/gad.17280911
Transcriptional regulation of xenobiotic detoxification in Drosophila
Abstract
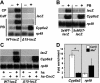
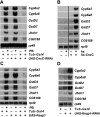
Living organisms, from bacteria to humans, display a coordinated transcriptional response to xenobiotic exposure, inducing enzymes and transporters that facilitate detoxification. Several transcription factors have been identified in vertebrates that contribute to this regulatory response. In contrast, little is known about this pathway in insects. Here we show that the Drosophila Nrf2 (NF-E2-related factor 2) ortholog CncC (cap 'n' collar isoform-C) is a central regulator of xenobiotic detoxification responses. A binding site for CncC and its heterodimer partner Maf (muscle aponeurosis fibromatosis) is sufficient and necessary for robust transcriptional responses to three xenobiotic compounds: phenobarbital (PB), chlorpromazine, and caffeine. Genetic manipulations that alter the levels of CncC or its negative regulator, Keap1 (Kelch-like ECH-associated protein 1), lead to predictable changes in xenobiotic-inducible gene expression. Transcriptional profiling studies reveal that more than half of the genes regulated by PB are also controlled by CncC. Consistent with these effects on detoxification gene expression, activation of the CncC/Keap1 pathway in Drosophila is sufficient to confer resistance to the lethal effects of the pesticide malathion. These studies establish a molecular mechanism for the regulation of xenobiotic detoxification in Drosophila and have implications for controlling insect populations and the spread of insect-borne human diseases.
Figures




References
-
- Abdollahi M, Ranjbar A, Shadnia S, Nikfar S, Rezaiee A 2004. Pesticides and oxidative stress: a review. Med Sci Monit 10: RA141–RA147 - PubMed
-
- Bhat R, Bresnick E 1997. Glycine N-methyltransferase is an example of functional diversity. Role as a polycyclic aromatic hydrocarbon-binding receptor. J Biol Chem 272: 21221–21226 - PubMed
-
- Brown RP, McDonnell CM, Berenbaum MR, Schuler MA 2005. Regulation of an insect cytochrome P450 monooxygenase gene (CYP6B1) by aryl hydrocarbon and xanthotoxin response cascades. Gene 358: 39–52 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
