Information theory and the ethylene genetic network
- PMID: 21897127
- PMCID: PMC3256376
- DOI: 10.4161/psb.6.10.16424
Information theory and the ethylene genetic network
Abstract
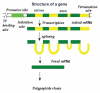
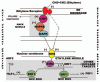
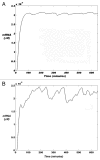
The original aim of the Information Theory (IT) was to solve a purely technical problem: to increase the performance of communication systems, which are constantly affected by interferences that diminish the quality of the transmitted information. That is, the theory deals only with the problem of transmitting with the maximal precision the symbols constituting a message. In Shannon's theory messages are characterized only by their probabilities, regardless of their value or meaning. As for its present day status, it is generally acknowledged that Information Theory has solid mathematical foundations and has fruitful strong links with Physics in both theoretical and experimental areas. However, many applications of Information Theory to Biology are limited to using it as a technical tool to analyze biopolymers, such as DNA, RNA or protein sequences. The main point of discussion about the applicability of IT to explain the information flow in biological systems is that in a classic communication channel, the symbols that conform the coded message are transmitted one by one in an independent form through a noisy communication channel, and noise can alter each of the symbols, distorting the message; in contrast, in a genetic communication channel the coded messages are not transmitted in the form of symbols but signaling cascades transmit them. Consequently, the information flow from the emitter to the effector is due to a series of coupled physicochemical processes that must ensure the accurate transmission of the message. In this review we discussed a novel proposal to overcome this difficulty, which consists of the modeling of gene expression with a stochastic approach that allows Shannon entropy (H) to be directly used to measure the amount of uncertainty that the genetic machinery has in relation to the correct decoding of a message transmitted into the nucleus by a signaling pathway. From the value of H we can define a function I that measures the amount of information content in the input message that the cell's genetic machinery is processing during a given time interval. Furthermore, combining Information Theory with the frequency response analysis of dynamical systems we can examine the cell's genetic response to input signals with varying frequencies, amplitude and form, in order to determine if the cell can distinguish between different regimes of information flow from the environment. In the particular case of the ethylene signaling pathway, the amount of information managed by the root cell of Arabidopsis can be correlated with the frequency of the input signal. The ethylene signaling pathway cuts off very low and very high frequencies, allowing a window of frequency response in which the nucleus reads the incoming message as a varying input. Outside of this window the nucleus reads the input message as an approximately non-varying one. This frequency response analysis is also useful to estimate the rate of information transfer during the transport of each new ERF1 molecule into the nucleus. Additionally, application of Information Theory to analysis of the flow of information in the ethylene signaling pathway provides a deeper insight in the form in which the transition between auxin and ethylene hormonal activity occurs during a circadian cycle. An ambitious goal for the future would be to use Information Theory as a theoretical foundation for a suitable model of the information flow that runs at each level and through all levels of biological organization.
Figures






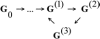
References
-
- Shannon CE. A mathematical theory of communication. Bell Syst Tech J. 1948;27:379–423.
-
- Weaver W. Recent contributions to the mathematical theory of communication. In: Shannon CE, Weaver W, editors. Mathematical theory of communication. Urbana, IL: University of Illinois Press; 1949.
-
- Johnson HA. Information Theory in Biology after 18 Years. Science. 1970;168:1545–1550. - PubMed
-
- Quastler H. Information Theory in Biology. Urbana, IL: University of Illinois Press; 1953.
-
- Shannon FF. Information theory and molecular biology. J Interdiscipl Math. 2009;12:41–87.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
