Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python
- PMID: 21897815
- PMCID: PMC3159964
- DOI: 10.3389/fninf.2011.00013
Nipype: a flexible, lightweight and extensible neuroimaging data processing framework in python
Abstract
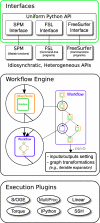
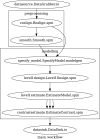
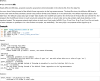
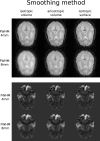
Current neuroimaging software offer users an incredible opportunity to analyze their data in different ways, with different underlying assumptions. Several sophisticated software packages (e.g., AFNI, BrainVoyager, FSL, FreeSurfer, Nipy, R, SPM) are used to process and analyze large and often diverse (highly multi-dimensional) data. However, this heterogeneous collection of specialized applications creates several issues that hinder replicable, efficient, and optimal use of neuroimaging analysis approaches: (1) No uniform access to neuroimaging analysis software and usage information; (2) No framework for comparative algorithm development and dissemination; (3) Personnel turnover in laboratories often limits methodological continuity and training new personnel takes time; (4) Neuroimaging software packages do not address computational efficiency; and (5) Methods sections in journal articles are inadequate for reproducing results. To address these issues, we present Nipype (Neuroimaging in Python: Pipelines and Interfaces; http://nipy.org/nipype), an open-source, community-developed, software package, and scriptable library. Nipype solves the issues by providing Interfaces to existing neuroimaging software with uniform usage semantics and by facilitating interaction between these packages using Workflows. Nipype provides an environment that encourages interactive exploration of algorithms, eases the design of Workflows within and between packages, allows rapid comparative development of algorithms and reduces the learning curve necessary to use different packages. Nipype supports both local and remote execution on multi-core machines and clusters, without additional scripting. Nipype is Berkeley Software Distribution licensed, allowing anyone unrestricted usage. An open, community-driven development philosophy allows the software to quickly adapt and address the varied needs of the evolving neuroimaging community, especially in the context of increasing demand for reproducible research.
Keywords: Python; data processing; neuroimaging; pipeline; reproducible research; workflow.
Figures





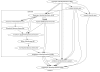
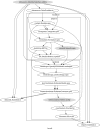
References
-
- Callahan S. P., Freire J., Santos E., Scheidegger C. E., Silva C. T., Vo H. T. (2006). “VisTrails: visualization meets data management,” in Proceedings of the 2006 ACM SIGMOD International Conference on Management of Data, Chicago, IL, 745–747
-
- Churchill N. W., Oder A., Abdi H., Tam F., Lee W., Thomas C., Ween J. E., Graham S. J., Strother S. C. (2011). Optimizing preprocessing and analysis pipelines for single-subject fMRI. I. Standard temporal motion and physiological noise correction methods. Hum. Brain Mapp. [Epub ahead of print].10.1002/hbm.21238 - DOI - PMC - PubMed
-
- Cointepas Y., Mangin J., Garnero L., Poline J. (2001). BrainVISA: software platform for visualization and analysis of multi-modality brain data. Neuroimage 13, 98.10.1016/S1053-8119(01)91441-7 - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

