Global distribution of Polaromonas phylotypes--evidence for a highly successful dispersal capacity
- PMID: 21897856
- PMCID: PMC3163589
- DOI: 10.1371/journal.pone.0023742
Global distribution of Polaromonas phylotypes--evidence for a highly successful dispersal capacity
Abstract
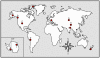
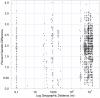
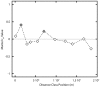
Bacteria from the genus Polaromonas are dominant phylotypes in clone libraries and culture collections from polar and high-elevation environments. Although Polaromonas has been found on six continents, we do not know if the same phylotypes exist in all locations or if they exhibit genetic isolation by distance patterns. To examine their biogeographic distribution, we analyzed all available, long-read 16S rRNA gene sequences of Polaromonas phylotypes from glacial and periglacial environments across the globe. Using genetic isolation by geographic distance analyses, including Mantel tests and Mantel correlograms, we found that Polaromonas phylotypes are globally distributed showing weak isolation by distance patterns at global scales. More focused analyses using discrete, equally sampled distances classes, revealed that only two distance classes (out of 12 total) showed significant spatial structuring. Overall, our analyses show that most Polaromonas phylotypes are truly globally distributed, but that some, as yet unknown, environmental variable may be selecting for unique phylotypes at a minority of our global sites. Analyses of aerobiological and genomic data suggest that Polaromonas phylotypes are globally distributed as dormant cells through high-elevation air currents; Polaromonas phylotypes are common in air and snow samples from high altitudes, and a glacial-ice metagenome and the two sequenced Polaromonas genomes contain the gene hipA, suggesting that Polaromonas can form dormant cells.
Conflict of interest statement
Figures

Similar articles
-
Bacterial community structure on two alpine debris-covered glaciers and biogeography of Polaromonas phylotypes.ISME J. 2013 Aug;7(8):1483-92. doi: 10.1038/ismej.2013.48. Epub 2013 Mar 28. ISME J. 2013. PMID: 23535918 Free PMC article.
-
Evidence of adaptation, niche separation and microevolution within the genus Polaromonas on Arctic and Antarctic glacial surfaces.Extremophiles. 2016 Jul;20(4):403-13. doi: 10.1007/s00792-016-0831-0. Epub 2016 Apr 20. Extremophiles. 2016. PMID: 27097637 Free PMC article.
-
Comparative genomics reveals the high diversity and adaptation strategies of Polaromonas from polar environments.BMC Genomics. 2025 Mar 14;26(1):248. doi: 10.1186/s12864-025-11410-6. BMC Genomics. 2025. PMID: 40087550 Free PMC article.
-
Polaromonas glacialis sp. nov. and Polaromonas cryoconiti sp. nov., isolated from alpine glacier cryoconite.Int J Syst Evol Microbiol. 2012 Nov;62(Pt 11):2662-2668. doi: 10.1099/ijs.0.037556-0. Epub 2011 Dec 23. Int J Syst Evol Microbiol. 2012. PMID: 22199222
-
The disappearing periglacial ecosystem atop Mt. Kilimanjaro supports both cosmopolitan and endemic microbial communities.Sci Rep. 2019 Jul 23;9(1):10676. doi: 10.1038/s41598-019-46521-0. Sci Rep. 2019. PMID: 31337772 Free PMC article.
Cited by
-
Linking the composition of cryoconite prokaryotic communities in the Arctic, Antarctic, and Central Caucasus with their chemical characteristics.Sci Rep. 2024 Jul 9;14(1):15838. doi: 10.1038/s41598-024-64452-3. Sci Rep. 2024. PMID: 38982048 Free PMC article.
-
The Microbial Olympics 2016.Nat Microbiol. 2016 Jul 26;1(8):16122. doi: 10.1038/nmicrobiol.2016.122. Nat Microbiol. 2016. PMID: 27573121 Free PMC article.
-
Microbiota entrapped in recently-formed ice: Paradana Ice Cave, Slovenia.Sci Rep. 2021 Jan 21;11(1):1993. doi: 10.1038/s41598-021-81528-6. Sci Rep. 2021. PMID: 33479448 Free PMC article.
-
Metagenomic insights into microbial community structure and metabolism in alpine permafrost on the Tibetan Plateau.Nat Commun. 2024 Jul 14;15(1):5920. doi: 10.1038/s41467-024-50276-2. Nat Commun. 2024. PMID: 39004662 Free PMC article.
-
Spatial patterns of benthic biofilm diversity among streams draining proglacial floodplains.Front Microbiol. 2022 Aug 8;13:948165. doi: 10.3389/fmicb.2022.948165. eCollection 2022. Front Microbiol. 2022. PMID: 36003939 Free PMC article.
References
-
- Bell T, Ager D, Song J-I, Newman JA, Thompson IP, et al. Larger islands house more bacterial taxa. Science. 2005;308:1884–1884. - PubMed
-
- Martiny JBH, Bohannan BJM, Brown JH, Colwell RK, Fuhrman JA, et al. Microbial biogeography: putting microorganisms on the map. Nat Rev Microbiol. 2006;4:102–112. - PubMed
-
- King AJ, Freeman KR, McCormick KF, Lynch RC, Lozupone C, et al. Biogeography and habitat modelling of high-alpine bacteria. Nat Commun. 2010;1:53. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

