KIR genotypic diversity can track ancestries in heterogeneous populations: a potential confounder for disease association studies
- PMID: 21898189
- PMCID: PMC4143378
- DOI: 10.1007/s00251-011-0569-x
KIR genotypic diversity can track ancestries in heterogeneous populations: a potential confounder for disease association studies
Abstract
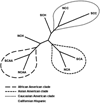
Killer cell immunoglobulin-like receptors (KIR) are encoded by highly polymorphic genes that regulate the activation of natural killer (NK) cells and other lymphocyte subsets and likely play key roles in innate and adaptive immunity. Association studies increasingly implicate KIR in disease predisposition and outcome but could be confounded by unknown KIR genetic structure in heterogeneous populations. To examine this, we characterized the diversity of 16 KIR genes in 712 Northern Californians (NC) stratified by self-assigned ethnicities and compared the profiles of KIR polymorphism with other US and global populations using a reference database. Sixty-eight distinct KIR genotypes were characterized: 58 in 457 Caucasians (NCC), 17 in 47 African Americans (NCAA), 21 in 80 Asians (NCA), 20 in 74 Hispanics (NCH), and 18 in 54 "other" ethnicities (NCO). KIR genotype patterns and frequencies in the 4 defined ethnicities were compared with each other and with 34 global populations by phylogenetic analysis. Although there were no population-specific genotypes, the KIR genotype frequency patterns faithfully traced the ancestry of NCC, NCAA, and NCA but not of NCH whose ancestries are known to be more heterogeneous. KIR genotype frequencies can therefore track ethnic ancestries in modern urban populations. Our data emphasize the importance of selecting ethnically matched controls in KIR-based studies to avert spurious associations.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the organizations that funded the research.
Figures


Similar articles
-
Predominance of B haplotype associated KIR genes in Tamil Speaking Dravidians.Hum Immunol. 2015 May;76(5):344-7. doi: 10.1016/j.humimm.2015.03.004. Epub 2015 Apr 1. Hum Immunol. 2015. PMID: 25842054
-
Distribution of KIR genes and KIR2DS4 gene variants in two Mexican Mestizo populations.Hum Immunol. 2017 Oct;78(10):614-620. doi: 10.1016/j.humimm.2017.07.010. Epub 2017 Jul 19. Hum Immunol. 2017. PMID: 28734803
-
Killer cell immunoglobulin-like receptor (KIR) locus profiles in the Tunisian population.Hum Immunol. 2015 May;76(5):355-61. doi: 10.1016/j.humimm.2015.03.002. Epub 2015 Mar 19. Hum Immunol. 2015. PMID: 25797201
-
Study of the KIR gene profiles and analysis of the phylogenetic relationships of Rajbanshi population of West Bengal, India.Hum Immunol. 2013 May;74(5):673-80. doi: 10.1016/j.humimm.2013.01.007. Epub 2013 Jan 24. Hum Immunol. 2013. PMID: 23354323
-
Autoimmune rheumatic diseases and their association with killer immunoglobulin-like receptor genes.Rev Bras Reumatol. 2011 Jul-Aug;51(4):351-6, 362-4. Rev Bras Reumatol. 2011. PMID: 21779711 Review. English, Portuguese.
Cited by
-
HLA-A29 and Birdshot Uveitis: Further Down the Rabbit Hole.Front Immunol. 2020 Nov 11;11:599558. doi: 10.3389/fimmu.2020.599558. eCollection 2020. Front Immunol. 2020. PMID: 33262772 Free PMC article. Review.
References
-
- Barbujani G, Colonna V. Human genome diversity: frequently asked questions. Trends Genet. 2010;26:285–295. - PubMed
-
- Bingham J, Sudarsanam S. Visualizing large hierarchical clusters in hyperbolic space. Bioinformatics. 2000;16:660–661. - PubMed
-
- Carrington M, Wang S, Martin MP, Gao X, Schiffman M, Cheng J, Herrero R, Rodriguez AC, Kurman R, Mortel R, Schwartz P, Glass A, Hildesheim A. Hierarchy of resistance to cervical neoplasia mediated by combinations of killer immunoglobulin-like receptor and human leukocyte antigen loci. J Exp Med. 2005;201:1069–1075. - PMC - PubMed
-
- Cavalli-Sforza LL, Menozzi P, Piazza A. The History and Geography of Human Genes. Princeton University Press; New Jersey: 1994. Europe; pp. 270–276.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

