RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection
- PMID: 21900162
- PMCID: PMC3209292
- DOI: 10.1128/JVI.05360-11
RelA Ser276 phosphorylation-coupled Lys310 acetylation controls transcriptional elongation of inflammatory cytokines in respiratory syncytial virus infection
Abstract
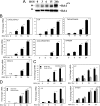
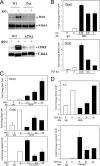
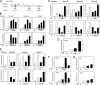
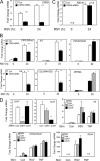
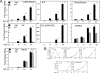
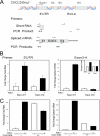
Respiratory syncytial virus (RSV) is a negative-sense single-stranded RNA virus responsible for lower respiratory tract infections (LRTIs) in humans. In experimental models of RSV LRTI, the actions of the nuclear factor κB (NF-κB) transcription factor mediate inflammation and pathology. We have shown that RSV replication induces a mitogen-and-stress-related kinase 1 (MSK-1) pathway that activates NF-κB RelA transcriptional activity by a process involving serine phosphorylation at serine (Ser) residue 276. In this study, we examined the mechanism by which phospho-Ser276 RelA mediates expression of the NF-κB-dependent gene network. RelA-deficient mouse embryonic fibroblasts (MEFs) complemented with the RelA Ser276Ala mutant are deficient in CXCL2/Groβ, KC, and interleukin-6 (IL-6) expression, but NFKBIA/IκBα is preserved. We show that RSV-induced RelA Ser276 phosphorylation is required for acetylation at Lys310, an event required for transcriptional activity and stable association of RelA with the activated positive transcriptional elongation factor (PTEF-b) complex proteins, bromodomain 4 (Brd4), and cyclin-dependent kinase 9 (CDK9). In contrast to gene loading pattern of PTEF-b proteins produced by tumor necrosis factor (TNF) stimulation, RSV induces their initial clearance followed by partial reaccumulation coincident with RelA recruitment. The RSV-induced binding patterns of the CDK9 substrate, phospho-Ser2 RNA polymerase (Pol) II, follows a similar pattern of clearance and downstream gene reaccumulation. The functional role of CDK9 was examined using CDK9 small interfering RNA (siRNA) and CDK inhibitors, where RSV-induced NF-κB-dependent gene expression was significantly inhibited. Finally, although RSV induces a transition from short transcripts to fully spliced mRNA in wild-type RelA (RelA WT)-expressing cells, this transition is not seen in cells expressing RelA Ser276Ala. We conclude that RelA Ser276 phosphorylation mediates RelA acetylation, Brd4/CDK9 association, and activation of downstream inflammatory genes by transcriptional elongation in RSV infection.
Figures




Similar articles
-
BRD4 Couples NF-κB/RelA with Airway Inflammation and the IRF-RIG-I Amplification Loop in Respiratory Syncytial Virus Infection.J Virol. 2017 Feb 28;91(6):e00007-17. doi: 10.1128/JVI.00007-17. Print 2017 Mar 15. J Virol. 2017. PMID: 28077651 Free PMC article.
-
Central Role of the NF-κB Pathway in the Scgb1a1-Expressing Epithelium in Mediating Respiratory Syncytial Virus-Induced Airway Inflammation.J Virol. 2018 May 14;92(11):e00441-18. doi: 10.1128/JVI.00441-18. Print 2018 Jun 1. J Virol. 2018. PMID: 29593031 Free PMC article.
-
RelA Ser276 phosphorylation is required for activation of a subset of NF-kappaB-dependent genes by recruiting cyclin-dependent kinase 9/cyclin T1 complexes.Mol Cell Biol. 2008 Jun;28(11):3623-38. doi: 10.1128/MCB.01152-07. Epub 2008 Mar 24. Mol Cell Biol. 2008. PMID: 18362169 Free PMC article.
-
Respiratory syncytial virus infection induces a reactive oxygen species-MSK1-phospho-Ser-276 RelA pathway required for cytokine expression.J Virol. 2009 Oct;83(20):10605-15. doi: 10.1128/JVI.01090-09. Epub 2009 Aug 12. J Virol. 2009. PMID: 19706715 Free PMC article.
-
RSV Reprograms the CDK9•BRD4 Chromatin Remodeling Complex to Couple Innate Inflammation to Airway Remodeling.Viruses. 2020 Apr 22;12(4):472. doi: 10.3390/v12040472. Viruses. 2020. PMID: 32331282 Free PMC article. Review.
Cited by
-
NF-κB activation by equine arteritis virus is MyD88 dependent and promotes viral replication.Arch Virol. 2013 Mar;158(3):701-5. doi: 10.1007/s00705-012-1515-4. Epub 2012 Nov 15. Arch Virol. 2013. PMID: 23151818 Free PMC article.
-
Targeting inducible epigenetic reprogramming pathways in chronic airway remodeling.Drugs Context. 2019 Oct 23;8:2019-8-3. doi: 10.7573/dic.2019-8-3. eCollection 2019. Drugs Context. 2019. PMID: 31692901 Free PMC article. Review.
-
Inhibition of BET Protein Function Suppressed the Overactivation of the Canonical NF-κB Signaling Pathway in 6-OHDA-Lesioned Rat Model of Levodopa-Induced Dyskinesia.Front Neurosci. 2022 Jun 21;16:896322. doi: 10.3389/fnins.2022.896322. eCollection 2022. Front Neurosci. 2022. PMID: 35801173 Free PMC article.
-
BET Family Protein BRD4: An Emerging Actor in NFκB Signaling in Inflammation and Cancer.Biomedicines. 2018 Feb 6;6(1):16. doi: 10.3390/biomedicines6010016. Biomedicines. 2018. PMID: 29415456 Free PMC article. Review.
-
Respiratory Syncytial Virus Infection Triggers Epithelial HMGB1 Release as a Damage-Associated Molecular Pattern Promoting a Monocytic Inflammatory Response.J Virol. 2016 Oct 14;90(21):9618-9631. doi: 10.1128/JVI.01279-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27535058 Free PMC article.
References
-
- Anrather J., Racchumi G., Iadecola C. 2005. cis-acting element-specific transcriptional activity of differentially phosphorylated nuclear factor-κB. J. Biol. Chem. 280: 244–252 - PubMed
-
- Barnes P. J., Karin M. 1997. Nuclear factor-kappaB: a pivotal transcription factor in chronic inflammatory diseases. N. Engl. J. Med. 336: 1066–1071 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

