Membrane-targeted WAVE mediates photoreceptor axon targeting in the absence of the WAVE complex in Drosophila
- PMID: 21900504
- PMCID: PMC3204070
- DOI: 10.1091/mbc.E11-02-0121
Membrane-targeted WAVE mediates photoreceptor axon targeting in the absence of the WAVE complex in Drosophila
Abstract
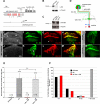
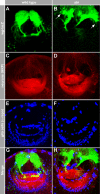
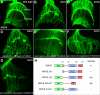
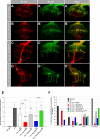
A tight spatial-temporal coordination of F-actin dynamics is crucial for a large variety of cellular processes that shape cells. The Abelson interactor (Abi) has a conserved role in Arp2/3-dependent actin polymerization, regulating Wiskott-Aldrich syndrome protein (WASP) and WASP family verprolin-homologous protein (WAVE). In this paper, we report that Abi exerts nonautonomous control of photoreceptor axon targeting in the Drosophila visual system through WAVE. In abi mutants, WAVE is unstable but restored by reexpression of Abi, confirming that Abi controls the integrity of the WAVE complex in vivo. Remarkably, expression of a membrane-tethered WAVE protein rescues the axonal projection defects of abi mutants in the absence of the other subunits of the WAVE complex, whereas cytoplasmic WAVE only slightly affects the abi mutant phenotype. Thus complex formation not only stabilizes WAVE, but also provides further membrane-recruiting signals, resulting in an activation of WAVE.
Figures



Similar articles
-
Abp1 utilizes the Arp2/3 complex activator Scar/WAVE in bristle development.J Cell Sci. 2012 Aug 1;125(Pt 15):3578-89. doi: 10.1242/jcs.101451. Epub 2012 Mar 30. J Cell Sci. 2012. PMID: 22467854
-
Abi activates WASP to promote sensory organ development.Nat Cell Biol. 2005 Oct;7(10):977-84. doi: 10.1038/ncb1305. Epub 2005 Sep 11. Nat Cell Biol. 2005. PMID: 16155589
-
Kette regulates actin dynamics and genetically interacts with Wave and Wasp.Development. 2003 Sep;130(18):4427-37. doi: 10.1242/dev.00663. Development. 2003. PMID: 12900458
-
New insights into the regulation and cellular functions of the ARP2/3 complex.Nat Rev Mol Cell Biol. 2013 Jan;14(1):7-12. doi: 10.1038/nrm3492. Epub 2012 Dec 5. Nat Rev Mol Cell Biol. 2013. PMID: 23212475 Review.
-
The WASP-WAVE protein network: connecting the membrane to the cytoskeleton.Nat Rev Mol Cell Biol. 2007 Jan;8(1):37-48. doi: 10.1038/nrm2069. Nat Rev Mol Cell Biol. 2007. PMID: 17183359 Review.
Cited by
-
CYRI controls epidermal wound closure and cohesion of invasive border cell cluster in Drosophila.J Cell Biol. 2024 Dec 2;223(12):e202310153. doi: 10.1083/jcb.202310153. Epub 2024 Oct 25. J Cell Biol. 2024. PMID: 39453414 Free PMC article.
-
New insights into the molecular mechanisms of axon guidance receptor regulation and signaling.Curr Top Dev Biol. 2021;142:147-196. doi: 10.1016/bs.ctdb.2020.11.008. Epub 2021 Jan 18. Curr Top Dev Biol. 2021. PMID: 33706917 Free PMC article. Review.
-
Robo recruitment of the Wave regulatory complex plays an essential and conserved role in midline repulsion.Elife. 2021 Apr 12;10:e64474. doi: 10.7554/eLife.64474. Elife. 2021. PMID: 33843588 Free PMC article.
-
Ena/VASP proteins cooperate with the WAVE complex to regulate the actin cytoskeleton.Dev Cell. 2014 Sep 8;30(5):569-84. doi: 10.1016/j.devcel.2014.08.001. Dev Cell. 2014. PMID: 25203209 Free PMC article.
-
The conserved Pelado/ZSWIM8 protein regulates actin dynamics by promoting linear actin filament polymerization.Life Sci Alliance. 2022 Aug 8;5(12):e202201484. doi: 10.26508/lsa.202201484. Life Sci Alliance. 2022. PMID: 35940847 Free PMC article.
References
-
- Anton IM, Jones GE, Wandosell F, Geha R, Ramesh N. WASP-interacting protein (WIP): working in polymerisation and much more. Trends Cell Biol. 2007;17:555–562. - PubMed
-
- Bazigou E, Apitz H, Johansson J, Loren CE, Hirst EMA, Chen PL, Palmer RH, Salecker I. Anterograde jelly belly and Alk receptor tyrosine kinase signaling mediates retinal axon targeting in Drosophila. Cell. 2007;128:961–975. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

