The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity
- PMID: 21901088
- PMCID: PMC3161960
- DOI: 10.1371/journal.ppat.1002130
The plant pathogen Pseudomonas syringae pv. tomato is genetically monomorphic and under strong selection to evade tomato immunity
Abstract
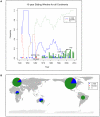
Recently, genome sequencing of many isolates of genetically monomorphic bacterial human pathogens has given new insights into pathogen microevolution and phylogeography. Here, we report a genome-based micro-evolutionary study of a bacterial plant pathogen, Pseudomonas syringae pv. tomato. Only 267 mutations were identified between five sequenced isolates in 3,543,009 nt of analyzed genome sequence, which suggests a recent evolutionary origin of this pathogen. Further analysis with genome-derived markers of 89 world-wide isolates showed that several genotypes exist in North America and in Europe indicating frequent pathogen movement between these world regions. Genome-derived markers and molecular analyses of key pathogen loci important for virulence and motility both suggest ongoing adaptation to the tomato host. A mutational hotspot was found in the type III-secreted effector gene hopM1. These mutations abolish the cell death triggering activity of the full-length protein indicating strong selection for loss of function of this effector, which was previously considered a virulence factor. Two non-synonymous mutations in the flagellin-encoding gene fliC allowed identifying a new microbe associated molecular pattern (MAMP) in a region distinct from the known MAMP flg22. Interestingly, the ancestral allele of this MAMP induces a stronger tomato immune response than the derived alleles. The ancestral allele has largely disappeared from today's Pto populations suggesting that flagellin-triggered immunity limits pathogen fitness even in highly virulent pathogens. An additional non-synonymous mutation was identified in flg22 in South American isolates. Therefore, MAMPs are more variable than expected differing even between otherwise almost identical isolates of the same pathogen strain.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

