Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration
- PMID: 21901095
- PMCID: PMC3161976
- DOI: 10.1371/journal.ppat.1002194
Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration
Abstract
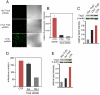
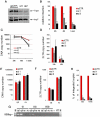
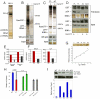
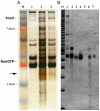
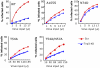
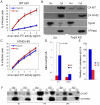
The HIV/AIDS pandemic is a major global health threat and understanding the detailed molecular mechanisms of HIV replication is critical for the development of novel therapeutics. To replicate, HIV-1 must access the nucleus of infected cells and integrate into host chromosomes, however little is known about the events occurring post-nuclear entry but before integration. Here we show that the karyopherin Transportin 3 (Tnp3) promotes HIV-1 integration in different cell types. Furthermore Tnp3 binds the viral capsid proteins and tRNAs incorporated into viral particles. Interaction between Tnp3, capsid and tRNAs is stronger in the presence of RanGTP, consistent with the possibility that Tnp3 is an export factor for these substrates. In agreement with this interpretation, we found that Tnp3 exports from the nuclei viral tRNAs in a RanGTP-dependent way. Tnp3 also binds and exports from the nuclei some species of cellular tRNAs with a defective 3'CCA end. Depletion of Tnp3 results in a re-distribution of HIV-1 capsid proteins between nucleus and cytoplasm however HIV-1 bearing the N74D mutation in capsid, which is insensitive to Tnp3 depletion, does not show nucleocytoplasmic redistribution of capsid proteins. We propose that Tnp3 promotes HIV-1 infection by displacing any capsid and tRNA that remain bound to the pre-integration complex after nuclear entry to facilitate integration. The results also provide evidence for a novel tRNA nucleocytoplasmic trafficking pathway in human cells.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Greber UF, Fassati A. Nuclear import of viral DNA genomes. Traffic. 2003;4:136–143. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

