A communal bacterial adhesin anchors biofilm and bystander cells to surfaces
- PMID: 21901100
- PMCID: PMC3161981
- DOI: 10.1371/journal.ppat.1002210
A communal bacterial adhesin anchors biofilm and bystander cells to surfaces
Abstract
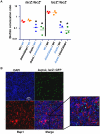
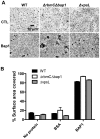
While the exopolysaccharide component of the biofilm matrix has been intensively studied, much less is known about matrix-associated proteins. To better understand the role of these proteins, we undertook a proteomic analysis of the V. cholerae biofilm matrix. Here we show that the two matrix-associated proteins, Bap1 and RbmA, perform distinct roles in the biofilm matrix. RbmA strengthens intercellular attachments. In contrast, Bap1 is concentrated on surfaces where it serves to anchor the biofilm and recruit cells not yet committed to the sessile lifestyle. This is the first example of a biofilm-derived, communally synthesized conditioning film that stabilizes the association of multilayer biofilms with a surface and facilitates recruitment of planktonic bystanders to the substratum. These studies define a novel paradigm for spatial and functional differentiation of proteins in the biofilm matrix and provide evidence for bacterial cooperation in maintenance and expansion of the multilayer biofilm.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
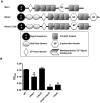
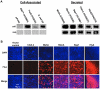

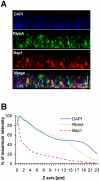
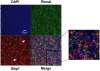



References
-
- Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 8:623–633. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

