Human butyrylcholinesterase-cocaine binding pathway and free energy profiles by molecular dynamics and potential of mean force simulations
- PMID: 21902185
- PMCID: PMC3179575
- DOI: 10.1021/jp2047807
Human butyrylcholinesterase-cocaine binding pathway and free energy profiles by molecular dynamics and potential of mean force simulations
Abstract
In the present study, we have performed combined molecular dynamics and potential of mean force (PMF) simulations to determine the enzyme-substrate (ES) binding pathway and the corresponding free energy profiles for wild-type butyrylcholinesterase (BChE) binding with (-)/(+)-cocaine and for the A328W/Y332G mutant binding with (-)-cocaine. According to the PMF simulations, for each ES binding system, the substrate first binds with the enzyme at a peripheral anionic site around the entrance of the active-site gorge to form the first ES complex (ES1-like) during the binding process. Further evolution from the ES1-like complex to the nonprereactive ES complex is nearly barrierless, with a free energy barrier lower than 1.0 kcal/mol. So, the nonprereactive ES binding process should be very fast. The rate-determining step of the entire ES binding process is the subsequent evolution from the nonprereactive ES complex to the prereactive ES complex. Further accounting for the entire ES binding process, the PMF-based simulations qualitatively reproduced the relative order of the experimentally derived binding free energies (ΔG(bind)), although the simulations systematically overestimated the magnitude of the binding affinity and systematically underestimated the differences between the ΔG(bind) values. The obtained structural and energetic insights into the entire ES binding process provide a valuable base for future rational design of high-activity mutants of BChE as candidates for an enzyme therapy for cocaine overdose and abuse.
© 2011 American Chemical Society
Figures

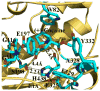



References
-
- Mendelson JH, Mello NK. New Eng J Med. 1996;334:965–972. - PubMed
-
- Sparenborg S, Vocci F, Zukin S. Drug Alcohol Depend. 1997;48:149–151. - PubMed
-
- Singh S. Chem Rev. 2000;100:925–1024. - PubMed
-
- Paula S, Tabet MR, Farr CD, Norman AB, Ball WJ., Jr J Med Chem. 2004;47:133–142. - PubMed
-
- Gorelick DA. Drug Alcohol Depend. 1997;48:159–165. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

