Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm
- PMID: 21903159
- PMCID: PMC3502750
- DOI: 10.1016/j.ygeno.2011.08.007
Design and coverage of high throughput genotyping arrays optimized for individuals of East Asian, African American, and Latino race/ethnicity using imputation and a novel hybrid SNP selection algorithm
Abstract
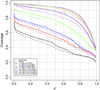
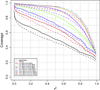
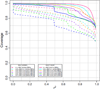
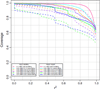
Four custom Axiom genotyping arrays were designed for a genome-wide association (GWA) study of 100,000 participants from the Kaiser Permanente Research Program on Genes, Environment and Health. The array optimized for individuals of European race/ethnicity was previously described. Here we detail the development of three additional microarrays optimized for individuals of East Asian, African American, and Latino race/ethnicity. For these arrays, we decreased redundancy of high-performing SNPs to increase SNP capacity. The East Asian array was designed using greedy pairwise SNP selection. However, removing SNPs from the target set based on imputation coverage is more efficient than pairwise tagging. Therefore, we developed a novel hybrid SNP selection method for the African American and Latino arrays utilizing rounds of greedy pairwise SNP selection, followed by removal from the target set of SNPs covered by imputation. The arrays provide excellent genome-wide coverage and are valuable additions for large-scale GWA studies.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures



References
-
- Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516–1517. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

