A brief survey of mRNA surveillance
- PMID: 21903397
- PMCID: PMC3205232
- DOI: 10.1016/j.tibs.2011.07.005
A brief survey of mRNA surveillance
Abstract
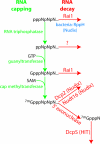
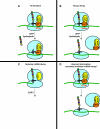
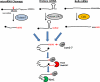
Defective mRNAs are degraded more rapidly than normal mRNAs in a process called mRNA surveillance. Eukaryotic cells use a variety of mechanisms to detect aberrations in mRNAs and a variety of enzymes to preferentially degrade them. Recent advances in the field of RNA surveillance have provided new information regarding how cells determine which mRNA species should be subject to destruction and novel mechanisms by which a cell tags an mRNA once such a decision has been reached. In this review, we highlight recent progress in our understanding of these processes.
Copyright © 2011 Elsevier Ltd. All rights reserved.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

