FLASH: fast length adjustment of short reads to improve genome assemblies
- PMID: 21903629
- PMCID: PMC3198573
- DOI: 10.1093/bioinformatics/btr507
FLASH: fast length adjustment of short reads to improve genome assemblies
Abstract
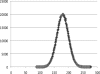
Motivation: Next-generation sequencing technologies generate very large numbers of short reads. Even with very deep genome coverage, short read lengths cause problems in de novo assemblies. The use of paired-end libraries with a fragment size shorter than twice the read length provides an opportunity to generate much longer reads by overlapping and merging read pairs before assembling a genome.
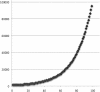
Results: We present FLASH, a fast computational tool to extend the length of short reads by overlapping paired-end reads from fragment libraries that are sufficiently short. We tested the correctness of the tool on one million simulated read pairs, and we then applied it as a pre-processor for genome assemblies of Illumina reads from the bacterium Staphylococcus aureus and human chromosome 14. FLASH correctly extended and merged reads >99% of the time on simulated reads with an error rate of <1%. With adequately set parameters, FLASH correctly merged reads over 90% of the time even when the reads contained up to 5% errors. When FLASH was used to extend reads prior to assembly, the resulting assemblies had substantially greater N50 lengths for both contigs and scaffolds.
Availability and implementation: The FLASH system is implemented in C and is freely available as open-source code at http://www.cbcb.umd.edu/software/flash.
Contact: t.magoc@gmail.com.
Figures
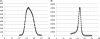

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

