Widespread signatures of recent selection linked to nucleosome positioning in the human lineage
- PMID: 21903742
- PMCID: PMC3205563
- DOI: 10.1101/gr.122275.111
Widespread signatures of recent selection linked to nucleosome positioning in the human lineage
Abstract
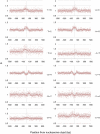
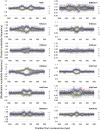
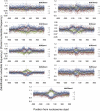
In this study we investigated the strengths and modes of selection associated with nucleosome positioning in the human lineage through the comparison of interspecies and intraspecies rates of divergence. We identify significant evidence for both positive and negative selection linked to human nucleosome positioning for the first time, implicating a widespread and important role for DNA sequence in the location of well-positioned nucleosomes. Selection appears to be acting on particular base substitutions to maintain optimum GC compositions in core and linker regions, with, e.g., unexpectedly elevated rates of C→T substitutions during recent human evolution at linker regions 60-90 bp from the nucleosome dyad but significant depletion of the same substitutions within nucleosome core regions. These patterns are strikingly consistent with the known relationships between genomic sequence composition and nucleosome assembly. By stratifying nucleosomes according to the GC content of their genomic neighborhood, we also show that the strength and direction of selection detected is dictated by local GC content. Intriguingly these signatures of selection are not restricted to nucleosomes in close proximity to exons, suggesting the correct positioning of nucleosomes is not only important in and around coding regions. This analysis provides strong evidence that the genomic sequences associated with nucleosomes are not evolving neutrally, and suggests that underlying DNA sequence is an important factor in nucleosome positioning. Recent signatures of selection linked to genomic features as ubiquitous as the nucleosome have important implications for human genome evolution and disease.
Figures




Similar articles
-
Nucleosomes shape DNA polymorphism and divergence.PLoS Genet. 2014 Jul 3;10(7):e1004457. doi: 10.1371/journal.pgen.1004457. eCollection 2014 Jul. PLoS Genet. 2014. PMID: 24991813 Free PMC article.
-
Evidence of selection for an accessible nucleosomal array in human.BMC Genomics. 2016 Jul 29;17:526. doi: 10.1186/s12864-016-2880-2. BMC Genomics. 2016. PMID: 27472913 Free PMC article.
-
Archaeal nucleosome positioning in vivo and in vitro is directed by primary sequence motifs.BMC Genomics. 2013 Jun 10;14:391. doi: 10.1186/1471-2164-14-391. BMC Genomics. 2013. PMID: 23758892 Free PMC article.
-
A structural perspective on the where, how, why, and what of nucleosome positioning.J Biomol Struct Dyn. 2010 Jun;27(6):803-20. doi: 10.1080/07391102.2010.10508585. J Biomol Struct Dyn. 2010. PMID: 20232935 Review.
-
Coupling between Sequence-Mediated Nucleosome Organization and Genome Evolution.Genes (Basel). 2021 Jun 1;12(6):851. doi: 10.3390/genes12060851. Genes (Basel). 2021. PMID: 34205881 Free PMC article. Review.
Cited by
-
Effects of DNA Methylation and Chromatin State on Rates of Molecular Evolution in Insects.G3 (Bethesda). 2015 Dec 4;6(2):357-63. doi: 10.1534/g3.115.023499. G3 (Bethesda). 2015. PMID: 26637432 Free PMC article.
-
Unusual mammalian usage of TGA stop codons reveals that sequence conservation need not imply purifying selection.PLoS Biol. 2022 May 12;20(5):e3001588. doi: 10.1371/journal.pbio.3001588. eCollection 2022 May. PLoS Biol. 2022. PMID: 35550630 Free PMC article.
-
Genomic approaches to DNA repair and mutagenesis.DNA Repair (Amst). 2015 Dec;36:146-155. doi: 10.1016/j.dnarep.2015.09.018. Epub 2015 Sep 15. DNA Repair (Amst). 2015. PMID: 26411877 Free PMC article. Review.
-
Predicting nucleosome binding motif set and analyzing their distributions around functional sites of human genes.Chromosome Res. 2012 Aug;20(6):685-98. doi: 10.1007/s10577-012-9305-0. Epub 2012 Jul 31. Chromosome Res. 2012. PMID: 22847645
-
Polyadenylation-related isoform switching in human evolution revealed by full-length transcript structure.Brief Bioinform. 2021 Nov 5;22(6):bbab157. doi: 10.1093/bib/bbab157. Brief Bioinform. 2021. PMID: 33973996 Free PMC article.
References
-
- Barski A, Cuddapah S, Cui K, Roh T-Y, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 - PubMed
-
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schubeler D 2010. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol 17: 894–900 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
