BrabA.11339.a: anomalous diffraction and ligand binding guide towards the elucidation of the function of a 'putative β-lactamase-like protein' from Brucella melitensis
- PMID: 21904058
- PMCID: PMC3169410
- DOI: 10.1107/S1744309111010220
BrabA.11339.a: anomalous diffraction and ligand binding guide towards the elucidation of the function of a 'putative β-lactamase-like protein' from Brucella melitensis
Abstract
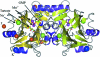
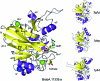
The crystal structure of a β-lactamase-like protein from Brucella melitensis was initially solved by SAD phasing from an in-house data set collected on a crystal soaked with iodide. A high-resolution data set was collected at a synchroton at the Se edge wavelength, which also provided an independent source of phasing using a small anomalous signal from metal ions in the active site. Comparisons of anomalous peak heights at various wavelengths allowed the identification of the active-site metal ions as manganese. In the native data set a partially occupied GMP could be identified. When co-crystallized with AMPPNP or GMPPNP, clear density for the hydrolyzed analogs was observed, providing hints to the function of the protein.
Figures


References
-
- Abendroth, J., Edwards, T. E., Staker, B. L., Myler, P. J. & Stewart, L. J. (2010). Northwest Crystallography Workshop, University of British Columbia, Canada.
-
- Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. (2003). Acta Cryst. D59, 2023–2030. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

