Structure of the cystathionine γ-synthase MetB from Mycobacterium ulcerans
- PMID: 21904066
- PMCID: PMC3169418
- DOI: 10.1107/S1744309111029575
Structure of the cystathionine γ-synthase MetB from Mycobacterium ulcerans
Abstract
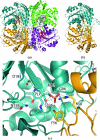
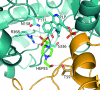
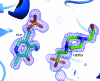
Cystathionine γ-synthase (CGS) is a transulfurication enzyme that catalyzes the first specific step in L-methionine biosynthesis by the reaction of O(4)-succinyl-L-homoserine and L-cysteine to produce L-cystathionine and succinate. Controlling the first step in L-methionine biosythesis, CGS is an excellent potential drug target. Mycobacterium ulcerans is a slow-growing mycobacterium that is the third most common form of mycobacterial infection, mainly infecting people in Africa, Australia and Southeast Asia. Infected patients display a variety of skin ailments ranging from indolent non-ulcerated lesions as well as ulcerated lesions. Here, the crystal structure of CGS from M. ulcerans covalently linked to the cofactor pyridoxal phosphate (PLP) is reported at 1.9 Å resolution. A second structure contains PLP as well as a highly ordered HEPES molecule in the active site acting as a pseudo-ligand. These results present the first structure of a CGS from a mycobacterium and allow comparison with other CGS enzymes. This is also the first structure reported from the pathogen M. ulcerans.
Figures

References
-
- Alexandrov, A., Dutta, K. & Pascal, S. M. (2001). Biotechniques, 30, 1194–1198. - PubMed
-
- Alexandrov, A., Vignali, M., LaCount, D. J., Quartley, E., de Vries, C., De Rosa, D., Babulski, J., Mitchell, S. F., Schoenfeld, L. W., Fields, S., Hol, W. G., Dumont, M. E., Phizicky, E. M. & Grayhack, E. J. (2004). Mol. Cell. Proteomics, 3, 934–938. - PubMed
-
- Amadasi, A., Bertoldi, M., Contestabile, R., Bettati, S., Cellini, B., di Salvo, M. L., Borri-Voltattorni, C., Bossa, F. & Mozzarelli, A. (2007). Curr. Med. Chem. 14, 1291–1324. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

