DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana
- PMID: 21904598
- PMCID: PMC3164146
- DOI: 10.1371/journal.pone.0023918
DELLA-induced early transcriptional changes during etiolated development in Arabidopsis thaliana
Abstract
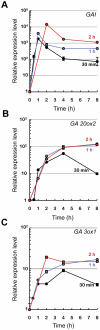
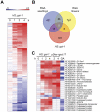
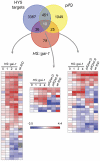
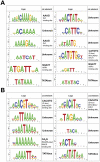
The hormones gibberellins (GAs) control a wide variety of processes in plants, including stress and developmental responses. This task largely relies on the activity of the DELLA proteins, nuclear-localized transcriptional regulators that do not seem to have DNA binding capacity. The identification of early target genes of DELLA action is key not only to understand how GAs regulate physiological responses, but also to get clues about the molecular mechanisms by which DELLAs regulate gene expression. Here, we have investigated the global, early transcriptional response triggered by the Arabidopsis DELLA protein GAI during skotomorphogenesis, a developmental program tightly regulated by GAs. Our results show that the induction of GAI activity has an almost immediate effect on gene expression. Although this transcriptional regulation is largely mediated by the PIFs and HY5 transcription factors based on target meta-analysis, additional evidence points to other transcription factors that would be directly involved in DELLA regulation of gene expression. First, we have identified cis elements recognized by Dofs and type-B ARRs among the sequences enriched in the promoters of GAI targets; and second, an enrichment in additional cis elements appeared when this analysis was extended to a dataset of early targets of the DELLA protein RGA: CArG boxes, bound by MADS-box proteins, and the E-box CACATG that links the activity of DELLAs to circadian transcriptional regulation. Finally, Gene Ontology analysis highlights the impact of DELLA regulation upon the homeostasis of the GA, auxin, and ethylene pathways, as well as upon pre-existing transcriptional networks.
Conflict of interest statement
Figures
Similar articles
-
GA-DELLA pathway is involved in regulation of nitrogen deficiency-induced anthocyanin accumulation.Plant Cell Rep. 2017 Apr;36(4):557-569. doi: 10.1007/s00299-017-2102-7. Epub 2017 Mar 8. Plant Cell Rep. 2017. PMID: 28275852
-
Evidence that the Arabidopsis nuclear gibberellin signalling protein GAI is not destabilised by gibberellin.Plant J. 2002 Dec;32(6):935-47. doi: 10.1046/j.1365-313x.2002.01478.x. Plant J. 2002. PMID: 12492836
-
DELLA-GAF1 complex is involved in tissue-specific expression and gibberellin feedback regulation of GA20ox1 in Arabidopsis.Plant Mol Biol. 2021 Oct;107(3):147-158. doi: 10.1007/s11103-021-01195-z. Epub 2021 Sep 25. Plant Mol Biol. 2021. PMID: 34562198
-
Genomic analysis of DELLA protein activity.Plant Cell Physiol. 2013 Aug;54(8):1229-37. doi: 10.1093/pcp/pct082. Epub 2013 Jun 18. Plant Cell Physiol. 2013. PMID: 23784221 Review.
-
DELLA and SCL3 balance gibberellin feedback regulation by utilizing INDETERMINATE DOMAIN proteins as transcriptional scaffolds.Plant Signal Behav. 2014;9(9):e29726. doi: 10.4161/psb.29726. Plant Signal Behav. 2014. PMID: 25763707 Free PMC article. Review.
Cited by
-
Sugar metabolism during pre- and post-fertilization events in plants under high temperature stress.Plant Cell Rep. 2022 Mar;41(3):655-673. doi: 10.1007/s00299-021-02795-1. Epub 2021 Oct 9. Plant Cell Rep. 2022. PMID: 34628530 Review.
-
DELLA proteins and their interacting RING Finger proteins repress gibberellin responses by binding to the promoters of a subset of gibberellin-responsive genes in Arabidopsis.Plant Cell. 2013 Mar;25(3):927-43. doi: 10.1105/tpc.112.108951. Epub 2013 Mar 12. Plant Cell. 2013. PMID: 23482857 Free PMC article.
-
Ectopic expression of specific GA2 oxidase mutants promotes yield and stress tolerance in rice.Plant Biotechnol J. 2017 Jul;15(7):850-864. doi: 10.1111/pbi.12681. Epub 2017 Mar 23. Plant Biotechnol J. 2017. PMID: 27998028 Free PMC article.
-
Characterization of Arabidopsis thaliana GCN2 kinase roles in seed germination and plant development.Plant Signal Behav. 2015;10(4):e992264. doi: 10.4161/15592324.2014.992264. Plant Signal Behav. 2015. PMID: 25912940 Free PMC article.
-
Arabidopsis DELLA and two HD-ZIP transcription factors regulate GA signaling in the epidermis through the L1 box cis-element.Plant Cell. 2014 Jul;26(7):2905-19. doi: 10.1105/tpc.114.127647. Epub 2014 Jul 2. Plant Cell. 2014. PMID: 24989044 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

