Mutation of L-2,3-diaminopropionic acid synthase genes blocks staphyloferrin B synthesis in Staphylococcus aureus
- PMID: 21906287
- PMCID: PMC3179956
- DOI: 10.1186/1471-2180-11-199
Mutation of L-2,3-diaminopropionic acid synthase genes blocks staphyloferrin B synthesis in Staphylococcus aureus
Abstract
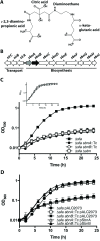
Background: Staphylococcus aureus synthesizes two siderophores, staphyloferrin A and staphyloferrin B, that promote iron-restricted growth. Previous work on the biosynthesis of staphyloferrin B has focused on the role of the synthetase enzymes, encoded from within the sbnA-I operon, which build the siderophore from the precursor molecules citrate, alpha-ketoglutarate and L-2,3-diaminopropionic acid. However, no information yet exists on several other enzymes, expressed from the biosynthetic cluster, that are thought to be involved in the synthesis of the precursors (or synthetase substrates) themselves.
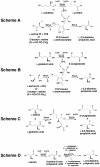
Results: Using mutants carrying insertions in sbnA and sbnB, we show that these two genes are essential for the synthesis of staphyloferrin B, and that supplementation of the growth medium with L-2,3-diaminopropionic acid can bypass the block in staphyloferrin B synthesis displayed by the mutants. Several mechanisms are proposed for how the enzymes SbnA, with similarity to cysteine synthase enzymes, and SbnB, with similarity to amino acid dehydrogenases and ornithine cyclodeaminases, function together in the synthesis of this unusual nonproteinogenic amino acid L-2,3-diaminopropionic acid.
Conclusions: Mutation of either sbnA or sbnB result in abrogation of synthesis of staphyloferrin B, a siderophore that contributes to iron-restricted growth of S. aureus. The loss of staphyloferrin B synthesis is due to an inability to synthesize the unusual amino acid L-2,3-diaminopropionic acid which is an important, iron-liganding component of the siderophore structure. It is proposed that SbnA and SbnB function together as an L-Dap synthase in the S. aureus cell.
Figures

Similar articles
-
Synthesis of L-2,3-diaminopropionic acid, a siderophore and antibiotic precursor.Chem Biol. 2014 Mar 20;21(3):379-88. doi: 10.1016/j.chembiol.2013.12.011. Epub 2014 Jan 30. Chem Biol. 2014. PMID: 24485762
-
Molecular characterization of staphyloferrin B biosynthesis in Staphylococcus aureus.Mol Microbiol. 2009 Nov;74(3):594-608. doi: 10.1111/j.1365-2958.2009.06880.x. Epub 2009 Sep 22. Mol Microbiol. 2009. PMID: 19775248
-
Deciphering the Substrate Specificity of SbnA, the Enzyme Catalyzing the First Step in Staphyloferrin B Biosynthesis.Biochemistry. 2016 Feb 16;55(6):927-39. doi: 10.1021/acs.biochem.5b01045. Epub 2016 Feb 3. Biochemistry. 2016. PMID: 26794841 Free PMC article.
-
Characterization of staphyloferrin A biosynthetic and transport mutants in Staphylococcus aureus.Mol Microbiol. 2009 May;72(4):947-63. doi: 10.1111/j.1365-2958.2009.06698.x. Epub 2009 Apr 14. Mol Microbiol. 2009. PMID: 19400778
-
β-N-Oxalyl-l-α,β-diaminopropionic Acid (β-ODAP) Content in Lathyrus sativus: The Integration of Nitrogen and Sulfur Metabolism through β-Cyanoalanine Synthase.Int J Mol Sci. 2017 Feb 28;18(3):526. doi: 10.3390/ijms18030526. Int J Mol Sci. 2017. PMID: 28264526 Free PMC article. Review.
Cited by
-
The Key Element Role of Metallophores in the Pathogenicity and Virulence of Staphylococcus aureus: A Review.Biology (Basel). 2022 Oct 18;11(10):1525. doi: 10.3390/biology11101525. Biology (Basel). 2022. PMID: 36290427 Free PMC article. Review.
-
The evolution of the ribosome and the genetic code.Life (Basel). 2014 May 20;4(2):227-49. doi: 10.3390/life4020227. Life (Basel). 2014. PMID: 25370196 Free PMC article.
-
A Heme-responsive Regulator Controls Synthesis of Staphyloferrin B in Staphylococcus aureus.J Biol Chem. 2016 Jan 1;291(1):29-40. doi: 10.1074/jbc.M115.696625. Epub 2015 Nov 3. J Biol Chem. 2016. PMID: 26534960 Free PMC article.
-
Characterization of an L-α,β-diaminopropionic acid polymer with comb-like structure isolated from a poly(ε-L-lysine)-producing Streptomyces sp.Appl Microbiol Biotechnol. 2021 Apr;105(8):3145-3157. doi: 10.1007/s00253-021-11257-3. Epub 2021 Apr 12. Appl Microbiol Biotechnol. 2021. PMID: 33846822
-
Iron Metabolism at the Interface between Host and Pathogen: From Nutritional Immunity to Antibacterial Development.Int J Mol Sci. 2020 Mar 20;21(6):2145. doi: 10.3390/ijms21062145. Int J Mol Sci. 2020. PMID: 32245010 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

