Systematic and quantitative assessment of the ubiquitin-modified proteome
- PMID: 21906983
- PMCID: PMC3200427
- DOI: 10.1016/j.molcel.2011.08.025
Systematic and quantitative assessment of the ubiquitin-modified proteome
Abstract
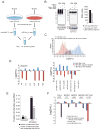
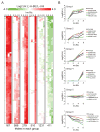
Despite the diverse biological pathways known to be regulated by ubiquitylation, global identification of substrates that are targeted for ubiquitylation has remained a challenge. To globally characterize the human ubiquitin-modified proteome (ubiquitinome), we utilized a monoclonal antibody that recognizes diglycine (diGly)-containing isopeptides following trypsin digestion. We identify ~19,000 diGly-modified lysine residues within ~5000 proteins. Using quantitative proteomics we monitored temporal changes in diGly site abundance in response to both proteasomal and translational inhibition, indicating both a dependence on ongoing translation to observe alterations in site abundance and distinct dynamics of individual modified lysines in response to proteasome inhibition. Further, we demonstrate that quantitative diGly proteomics can be utilized to identify substrates for cullin-RING ubiquitin ligases. Interrogation of the ubiquitinome allows for not only a quantitative assessment of alterations in protein homeostasis fidelity, but also identification of substrates for individual ubiquitin pathway enzymes.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures





References
-
- Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. - PubMed
-
- Benkirane M, Sardet C, Coux O. Lessons from interconnected ubiquitylation and acetylation of p53: think metastable networks. Biochem Soc Trans. 2010;38:98–103. - PubMed
-
- Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, Bates GP, Schulman H, Kopito RR. Global changes to the ubiquitin system in Huntington’s disease. Nature. 2007;448:704–708. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

