High-throughput protein purification and quality assessment for crystallization
- PMID: 21907284
- PMCID: PMC3690762
- DOI: 10.1016/j.ymeth.2011.07.010
High-throughput protein purification and quality assessment for crystallization
Abstract
The ultimate goal of structural biology is to understand the structural basis of proteins in cellular processes. In structural biology, the most critical issue is the availability of high-quality samples. "Structural biology-grade" proteins must be generated in the quantity and quality suitable for structure determination using X-ray crystallography or nuclear magnetic resonance (NMR) spectroscopy. The purification procedures must reproducibly yield homogeneous proteins or their derivatives containing marker atom(s) in milligram quantities. The choice of protein purification and handling procedures plays a critical role in obtaining high-quality protein samples. With structural genomics emphasizing a genome-based approach in understanding protein structure and function, a number of unique structures covering most of the protein folding space have been determined and new technologies with high efficiency have been developed. At the Midwest Center for Structural Genomics (MCSG), we have developed semi-automated protocols for high-throughput parallel protein expression and purification. A protein, expressed as a fusion with a cleavable affinity tag, is purified in two consecutive immobilized metal affinity chromatography (IMAC) steps: (i) the first step is an IMAC coupled with buffer-exchange, or size exclusion chromatography (IMAC-I), followed by the cleavage of the affinity tag using the highly specific Tobacco Etch Virus (TEV) protease; the second step is IMAC and buffer exchange (IMAC-II) to remove the cleaved tag and tagged TEV protease. These protocols have been implemented on multidimensional chromatography workstations and, as we have shown, many proteins can be successfully produced in large-scale. All methods and protocols used for purification, some developed by MCSG, others adopted and integrated into the MCSG purification pipeline and more recently the Center for Structural Genomics of Infectious Diseases (CSGID) purification pipeline, are discussed in this chapter.
Copyright © 2011 Elsevier Inc. All rights reserved.
Figures


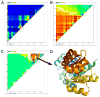


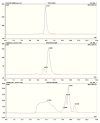
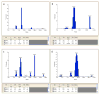

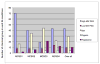
References
-
- Giglio MG, Collmer CW, Lomax J, Ireland A. Applying the Gene Ontology in microbial annotation. Trends Microbiol. 2009;17:262–268. - PubMed
-
- Pestka S. Production and analysis of proteins by recombinant DNA technology. Bioprocess Technol. 1990;7:235–265. - PubMed
-
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41:207–234. - PubMed
-
- Austin BP, Nallamsetty S, Waugh DS. Hexahistidine-tagged maltose-binding protein as a fusion partner for the production of soluble recombinant proteins in Escherichia coli. Methods Mol Biol. 2009;498:157–172. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

