Variability of Escherichia coli O157 strain survival in manure-amended soil in relation to strain origin, virulence profile, and carbon nutrition profile
- PMID: 21908630
- PMCID: PMC3208984
- DOI: 10.1128/AEM.00745-11
Variability of Escherichia coli O157 strain survival in manure-amended soil in relation to strain origin, virulence profile, and carbon nutrition profile
Abstract
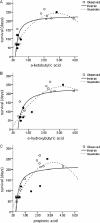
The variation in manure-amended soil survival capability among 18 Escherichia coli O157 strains (8 animal, 1 food, and 9 human isolates) was studied using a single sandy soil sample and a single sample of cattle manure as the inoculum carrier. The virulence profiles of E. coli O157 strains were characterized by detection of virulence determinants (73 genes, 122 probes in duplicate) by using the Identibac E. coli genotyping DNA miniaturized microarray. Metabolic profiling was done by subjecting all strains to the Biolog phenotypic carbon microarray. Survival times (calculated as days needed to reach the detection limit using the Weibull model) ranged from 47 to 266 days (median, 120 days). Survival time was significantly higher for the group of human isolates (median, 211 days; minimum [min.], 71; maximum [max.], 266) compared to the group of animal isolates (median, 70 days; min., 47; max., 249) (P = 0.025). Although clustering of human versus animal strains was observed based on pulsed-field gel electrophoresis (PFGE) patterns, no relation between survival time and the presence of virulence genes was observed. Principal component analysis on the metabolic profiling data revealed distinct clustering of short- and long-surviving strains. The oxidization rate of propionic acid, α-ketobutyric acid, and α-hydroxybutyric acid was significantly higher for the long-surviving strains than for the short-surviving strains. The oxidative capacity of E. coli O157 strains may be regarded as a phenotypic marker for enhanced survival in manure-amended soil. The large variation observed in survival is of importance for risk assessment models.
Figures
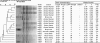


References
-
- Allerberger F., Sessitsch A. 2009. Incidence and microbiology of salad-borne disease. CAB Rev. 4:1–13
-
- Aluwong T., Kobo P. I., Abdullahi A. 2010. Volatile fatty acids production in ruminants and the role of monocarboxylate transporters: a review. J. Biotechnol. 9:6229–6232
-
- Aminul Islam M., Heuvelink A. E., Talukder K. A., de Boer E. 2006. Immunoconcentration of Shiga toxin-producing Escherichia coli O157 from animal faeces and raw meats by using Dynabeads anti-E. coli O157 and the VIDAS system. Int. J. Food Microbiol. 109:151–156 - PubMed
-
- Argenzio R. A., Southworth M., Stevens C. E. 1974. Sites of organic acid production and absorption in the equine gastrointestinal tract. Am. J. Physiol. 226:1043–1050 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

