Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma)
- PMID: 21908640
- PMCID: PMC3209139
- DOI: 10.1128/AEM.06059-11
Partial genome assembly for a candidate division OP11 single cell from an anoxic spring (Zodletone Spring, Oklahoma)
Abstract
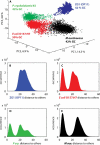
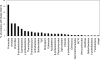
Members of candidate division OP11 are widely distributed in terrestrial and marine ecosystems, yet little information regarding their metabolic capabilities and ecological role within such habitats is currently available. Here, we report on the microfluidic isolation, multiple-displacement-amplification, pyrosequencing, and genomic analysis of a single cell (ZG1) belonging to candidate division OP11. Genome analysis of the ∼270-kb partial genome assembly obtained showed that it had no particular similarity to a specific phylum. Four hundred twenty-three open reading frames were identified, 46% of which had no function prediction. In-depth analysis revealed a heterotrophic lifestyle, with genes encoding endoglucanase, amylopullulanase, and laccase enzymes, suggesting a capacity for utilization of cellulose, starch, and, potentially, lignin, respectively. Genes encoding several glycolysis enzymes as well as formate utilization were identified, but no evidence for an electron transport chain was found. The presence of genes encoding various components of lipopolysaccharide biosynthesis indicates a Gram-negative bacterial cell wall. The partial genome also provides evidence for antibiotic resistance (β-lactamase, aminoglycoside phosphotransferase), as well as antibiotic production (bacteriocin) and extracellular bactericidal peptidases. Multiple mechanisms for stress response were identified, as were elements of type I and type IV secretion systems. Finally, housekeeping genes identified within the partial genome were used to demonstrate the OP11 affiliation of multiple hitherto unclassified genomic fragments from multiple database-deposited metagenomic data sets. These results provide the first glimpse into the lifestyle of a member of a ubiquitous, yet poorly understood bacterial candidate division.
Figures


References
-
- Beloqui A., et al. 2006. Novel polyphenol oxidase mined from a metagenome expression library of bovine rumen. J. Biol. Chem. 281:22933–22942 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

