Defense islands in bacterial and archaeal genomes and prediction of novel defense systems
- PMID: 21908672
- PMCID: PMC3194920
- DOI: 10.1128/JB.05535-11
Defense islands in bacterial and archaeal genomes and prediction of novel defense systems
Abstract
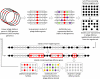
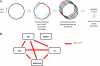
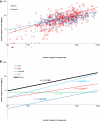
The arms race between cellular life forms and viruses is a major driving force of evolution. A substantial fraction of bacterial and archaeal genomes is dedicated to antivirus defense. We analyzed the distribution of defense genes and typical mobilome components (such as viral and transposon genes) in bacterial and archaeal genomes and demonstrated statistically significant clustering of antivirus defense systems and mobile genes and elements in genomic islands. The defense islands are enriched in putative operons and contain numerous overrepresented gene families. A detailed sequence analysis of the proteins encoded by genes in these families shows that many of them are diverged variants of known defense system components, whereas others show features, such as characteristic operonic organization, that are suggestive of novel defense systems. Thus, genomic islands provide abundant material for the experimental study of bacterial and archaeal antivirus defense. Except for the CRISPR-Cas systems, different classes of defense systems, in particular toxin-antitoxin and restriction-modification systems, show nonrandom clustering in defense islands. It remains unclear to what extent these associations reflect functional cooperation between different defense systems and to what extent the islands are genomic "sinks" that accumulate diverse nonessential genes, particularly those acquired via horizontal gene transfer. The characteristics of defense islands resemble those of mobilome islands. Defense and mobilome genes are nonrandomly associated in islands, suggesting nonadaptive evolution of the islands via a preferential attachment-like mechanism underpinned by the addictive properties of defense systems such as toxins-antitoxins and an important role of horizontal mobility in the evolution of these islands.
Figures






References
-
- Barrangou R., et al. 2007. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315:1709–1712 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

