Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar
- PMID: 21908685
- PMCID: PMC3252164
- DOI: 10.1104/pp.111.181354
Dissection of the transcriptional program regulating secondary wall biosynthesis during wood formation in poplar
Abstract
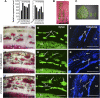
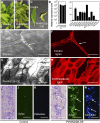
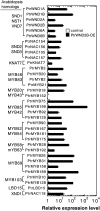
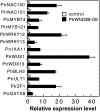
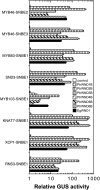
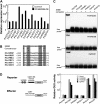
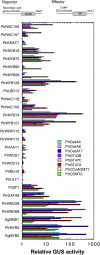
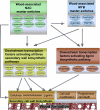
Wood biomass is mainly made of secondary cell walls; hence, elucidation of the molecular mechanisms underlying the transcriptional regulation of secondary wall biosynthesis during wood formation will be instrumental to design strategies for genetic improvement of wood biomass. Here, we provide direct evidence demonstrating that the poplar (Populus trichocarpa) wood-associated NAC domain transcription factors (PtrWNDs) are master switches activating a suite of downstream transcription factors, and together, they are involved in the coordinated regulation of secondary wall biosynthesis during wood formation. We show that transgenic poplar plants with dominant repression of PtrWNDs functions exhibit a drastic reduction in secondary wall thickening in woody cells, and those with PtrWND overexpression result in ectopic deposition of secondary walls. Analysis of PtrWND2B overexpressors revealed up-regulation of the expression of a number of wood-associated transcription factors, the promoters of which were also activated by PtrWND6B and the Eucalyptus EgWND1. Transactivation analysis and electrophoretic mobility shift assay demonstrated that PtrWNDs and EgWND1 activated gene expression through direct binding to the secondary wall NAC-binding elements, which are present in the promoters of several wood-associated transcription factors and a number of genes involved in secondary wall biosynthesis and modification. The WND-regulated transcription factors PtrNAC150, PtrNAC156, PtrNAC157, PtrMYB18, PtrMYB74, PtrMYB75, PtrMYB121, PtrMYB128, PtrZF1, and PtrGATA8 were able to activate the promoter activities of the biosynthetic genes for all three major wood components. Our study has uncovered that the WND master switches together with a battery of their downstream transcription factors form a transcriptional network controlling secondary wall biosynthesis during wood formation.
Figures





References
-
- Andersson-Gunnerås S, Mellerowicz EJ, Love J, Segerman B, Ohmiya Y, Coutinho PM, Nilsson P, Henrissat B, Moritz T, Sundberg B. (2006) Biosynthesis of cellulose-enriched tension wood in Populus: global analysis of transcripts and metabolites identifies biochemical and developmental regulators in secondary wall biosynthesis. Plant J 45: 144–165 - PubMed
-
- Burk DH, Zhong R, Morrison WH III, Ye Z-H. (2006) Disruption of cortical microtubules by overexpression of green fluorescent protein-tagged α-tubulin 6 causes a marked reduction in cell wall synthesis. J Integr Plant Biol 48: 85–98
-
- Carroll A, Somerville C. (2009) Cellulosic biofuels. Annu Rev Plant Biol 60: 165–182 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

