Light-dependent regulation of DEL1 is determined by the antagonistic action of E2Fb and E2Fc
- PMID: 21908689
- PMCID: PMC3252145
- DOI: 10.1104/pp.111.183384
Light-dependent regulation of DEL1 is determined by the antagonistic action of E2Fb and E2Fc
Erratum in
-
CORRECTION: Vol. 157: 1440-1451, 2011.Plant Physiol. 2017 Mar;173(3):1934-1935. doi: 10.1104/pp.17.00129. Plant Physiol. 2017. PMID: 28258122 Free PMC article. No abstract available.
Abstract
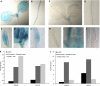
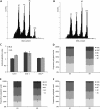
Endoreduplication represents a variation on the cell cycle in which multiple rounds of DNA replication occur without subsequent chromosome separation and cytokinesis, thereby increasing the cellular DNA content. It is known that the DNA ploidy level of cells is controlled by external stimuli such as light; however, limited knowledge is available on how environmental signals regulate the endoreduplication cycle at the molecular level. Previously, we had demonstrated that the conversion from a mitotic cell cycle into an endoreduplication cycle is controlled by the atypical E2F transcription factor, DP-E2F-LIKE1 (DEL1), that represses the endocycle onset. Here, the Arabidopsis (Arabidopsis thaliana) DEL1 gene was identified as a transcriptional target of the classical E2Fb and E2Fc transcription factors that antagonistically control its transcript levels through competition for a single E2F cis-acting binding site. In accordance with the reported opposite effects of light on the protein levels of E2Fb and E2Fc, DEL1 transcription depended on the light regime. Strikingly, modified DEL1 expression levels uncoupled the link between light and endoreduplication in hypocotyls, implying that DEL1 acts as a regulatory connection between endocycle control and the photomorphogenic response.
Figures





References
-
- Artlip TS, Madison JT, Setter TL. (1995) Water deficit in developing endosperm of maize: cell division and nuclear DNA endoreduplication. Plant Cell Environ 18: 1034–1040
-
- Berckmans B, De Veylder L. (2009) Transcriptional control of the cell cycle. Curr Opin Plant Biol 12: 599–605 - PubMed
-
- Boudolf V, Vlieghe K, Beemster GTS, Magyar Z, Torres Acosta JA, Maes S, Van Der Schueren E, Inzé D, De Veylder L. (2004) The plant-specific cyclin-dependent kinase CDKB1;1 and transcription factor E2Fa-DPa control the balance of mitotically dividing and endoreduplicating cells in Arabidopsis. Plant Cell 16: 2683–2692 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

