IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes
- PMID: 21909072
- PMCID: PMC3243593
- DOI: 10.1038/emboj.2011.325
IRF6 is a mediator of Notch pro-differentiation and tumour suppressive function in keratinocytes
Abstract
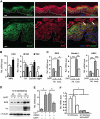
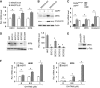
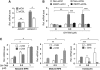
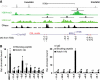
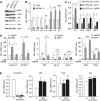
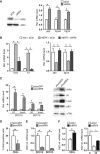
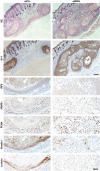
While the pro-differentiation and tumour suppressive functions of Notch signalling in keratinocytes are well established, the underlying mechanisms remain poorly understood. We report here that interferon regulatory factor 6 (IRF6), an IRF family member with an essential role in epidermal development, is induced in differentiation through a Notch-dependent mechanism and is a primary Notch target in keratinocytes and keratinocyte-derived SCC cells. Increased IRF6 expression contributes to the impact of Notch activation on growth/differentiation-related genes, while it is not required for induction of 'canonical' Notch targets like p21(WAF1/Cip1), Hes1 and Hey1. Down-modulation of IRF6 counteracts differentiation of primary human keratinocytes in vitro and in vivo, promoting ras-induced tumour formation. The clinical relevance of these findings is illustrated by the strikingly opposite pattern of expression of Notch1 and IRF6 versus epidermal growth factor receptor in a cohort of clinical SCCs, as a function of their grade of differentiation. Thus, IRF6 is a primary Notch target in keratinocytes, which contributes to the role of this pathway in differentiation and tumour suppression.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Artavanis-Tsakonas S, Rand MD, Lake RJ (1999) Notch signaling: cell fate control and signal integration in development. Science 284: 770–776 - PubMed
-
- Bailey CM, Khalkhali-Ellis Z, Kondo S, Margaryan NV, Seftor RE, Wheaton WW, Amir S, Pins MR, Schutte BC, Hendrix MJ (2005) Mammary serine protease inhibitor (Maspin) binds directly to interferon regulatory factor 6: identification of a novel serpin partnership. J Biol Chem 280: 34210–34217 - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

