Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1
- PMID: 21909073
- PMCID: PMC3243592
- DOI: 10.1038/emboj.2011.324
Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1
Abstract
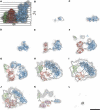
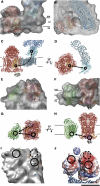
The respiratory chain in the inner mitochondrial membrane contains three large multi-enzyme complexes that together establish the proton gradient for ATP synthesis, and assemble into a supercomplex. A 19-Å 3D map of the 1.7-MDa amphipol-solubilized supercomplex I(1)III(2)IV(1) from bovine heart obtained by single-particle electron cryo-microscopy reveals an amphipol belt replacing the membrane lipid bilayer. A precise fit of the X-ray structures of complex I, the complex III dimer, and monomeric complex IV indicates distances of 13 nm between the ubiquinol-binding sites of complexes I and III, and of 10-11 nm between the cytochrome c binding sites of complexes III and IV. The arrangement of respiratory chain complexes suggests two possible pathways for efficient electron transfer through the supercomplex, of which the shorter branch through the complex III monomer proximal to complex I may be preferred.
Conflict of interest statement
J-LP holds patents on amphipols and their use. All other authors declare that they have no conflict of interest.
Figures






References
-
- Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA (2008) Respiratory active mitochondrial supercomplexes. Mol Cell 32: 529–539 - PubMed
-
- Ben-Shem A, Frolow F, Nelson N (2003) Crystal structure of plant photosystem I. Nature 426: 630–635 - PubMed
-
- Castellani M, Covian R, Kleinschroth T, Anderka O, Ludwig B, Trumpower BL (2010) Direct demonstration of half-of-the-sites reactivity in the dimeric cytochrome bc1 complex: enzyme with one inactive monomer is fully active but unable to activate the second ubiquinol oxidation site in response to ligand binding at the ubiquinone reduction site. J Biol Chem 285: 502–510 - PMC - PubMed
-
- Chance B, Williams GR (1955) Respiratory enzymes in oxidative phosphorylation. III. The steady state. J Biol Chem 217: 409–427 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

