Ligand-binding domain of an α7-nicotinic receptor chimera and its complex with agonist
- PMID: 21909087
- PMCID: PMC3489043
- DOI: 10.1038/nn.2908
Ligand-binding domain of an α7-nicotinic receptor chimera and its complex with agonist
Abstract
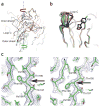
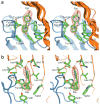
The α(7) acetylcholine receptor (AChR) mediates pre- and postsynaptic neurotransmission in the central nervous system and is a potential therapeutic target in neurodegenerative, neuropsychiatric and inflammatory disorders. We determined the crystal structure of the extracellular domain of a receptor chimera constructed from the human α(7) AChR and Lymnaea stagnalis acetylcholine binding protein (AChBP), which shares 64% sequence identity and 71% similarity with native α(7). We also determined the structure with bound epibatidine, a potent AChR agonist. Comparison of the structures revealed molecular rearrangements and interactions that mediate agonist recognition and early steps in signal transduction in α(7) AChRs. The structures further revealed a ring of negative charge within the central vestibule, poised to contribute to cation selectivity. Structure-guided mutational studies disclosed distinctive contributions to agonist recognition and signal transduction in α(7) AChRs. The structures provide a realistic template for structure-aided drug design and for defining structure-function relationships of α(7) AChRs.
© 2011 Nature America, Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing financial interests.
Figures


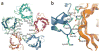
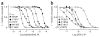
References
-
- Brejc K, et al. Crystal structure of an ACh-binding protein reveals the ligand-binding domain of nicotinic receptors. Nature. 2001;411:269–276. - PubMed
-
- Celie PH, et al. Nicotine and carbamylcholine binding to nicotinic acetylcholine receptors as studied in AChBP crystal structures. Neuron. 2004;41:907–914. - PubMed
-
- Unwin N. Refined structure of the nicotinic receptor at 4 Å resolution. J Mol Biol. 2005;346:967–989. - PubMed
-
- Dellisanti CD, Yao Y, Stroud JC, Wang ZZ, Chen L. Crystal structure of the extracellular domain of nAChR α1 bound to alpha-bungarotoxin at 1.94 Å resolution. Nat Neurosci. 2007;10:953–962. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

