Protonation of key acidic residues is critical for the K⁺-selectivity of the Na/K pump
- PMID: 21909093
- PMCID: PMC3190665
- DOI: 10.1038/nsmb.2113
Protonation of key acidic residues is critical for the K⁺-selectivity of the Na/K pump
Abstract
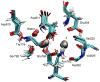
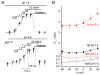
The sodium-potassium (Na/K) pump is a P-type ATPase that generates Na(+) and K(+) concentration gradients across the cell membrane. For each hydrolyzed ATP molecule, the pump extrudes three Na(+) and imports two K(+) by alternating between outward- and inward-facing conformations that preferentially bind K(+) or Na(+), respectively. Remarkably, the selective K(+) and Na(+) binding sites share several residues, and how the pump is able to achieve the selectivity required for the functional cycle is unclear. Here, free energy-perturbation molecular dynamics (FEP/MD) simulations based on the crystal structures of the Na/K pump in a K(+)-loaded state (E2·P(i)) reveal that protonation of the high-field acidic side chains involved in the binding sites is crucial to achieving the proper K(+) selectivity. This prediction is tested with electrophysiological experiments showing that the selectivity of the E2P state for K(+) over Na(+) is affected by extracellular pH.
Conflict of interest statement
Competing Interests
The authors declare that they have no competing financial interests.
Figures
References
-
- Jorgensen PL, Hakansson K+O, Karlish SJD. Structure and mechanism of Na+,K+-ATPase: Functional sites and their interactions. Annu Rev Physiol. 2003;65:817–849. - PubMed
-
- Post RL, Sen AK, Rosenthal AS. A phosphorylated intermediate in adenosine triphosphate-dependent sodium and potassium transport across K+idney membranes. J Biol Chem. 1965;240:1437–1445. - PubMed
-
- Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. - PubMed
-
- Albers RW. Biochemical aspects of active transport. Annu Rev Biochem. 1967;36:727–756. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

