Competition between ADAR and RNAi pathways for an extensive class of RNA targets
- PMID: 21909095
- PMCID: PMC3190075
- DOI: 10.1038/nsmb.2129
Competition between ADAR and RNAi pathways for an extensive class of RNA targets
Abstract
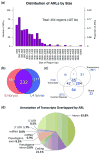
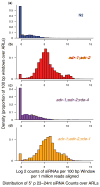
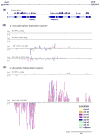
Adenosine deaminases that act on RNAs (ADARs) interact with double-stranded RNAs, deaminating adenosines to inosines. Previous studies of Caenorhabditis elegans indicated an antagonistic interaction between ADAR and RNAi machineries, with ADAR defects suppressed upon additional knockout of RNAi. This suggests a pool of common RNA substrates capable of engaging both pathways. To define and characterize such substrates, we examined small RNA and mRNA populations of ADAR mutants and identified a distinct set of loci from which RNAi-dependent short RNAs are markedly upregulated. At these same loci, we observed populations of multiply edited transcripts, supporting a specific role for ADARs in preventing access to the RNAi pathway for an extensive population of dsRNAs. Characterization of these loci revealed a substantial overlap with noncoding and intergenic regions, suggesting that the landscape of ADAR targets may extend beyond previously annotated classes of transcripts.
Figures




References
-
- Werry TD, Loiacono R, Sexton PM, Christopoulos A. RNA editing of the serotonin 5HT2C receptor and its effects on cell signalling, pharmacology and brain function. Pharmacol Ther. 2008;119:7–23. - PubMed
-
- Burns CM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. - PubMed
-
- Higuchi M, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNA-editing enzyme ADAR2. Nature. 2000;406:78–81. - PubMed
-
- Rula EY, Emeson RB. Mouse models to elucidate the functional roles of adenosine-to-inosine editing. Meth Enzymol. 2007;424:333–367. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

