Pancreatic mesenchyme regulates epithelial organogenesis throughout development
- PMID: 21909240
- PMCID: PMC3167782
- DOI: 10.1371/journal.pbio.1001143
Pancreatic mesenchyme regulates epithelial organogenesis throughout development
Abstract
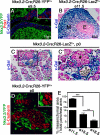
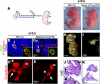
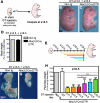
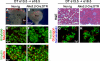
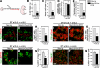
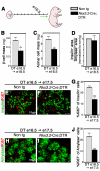
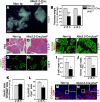
The developing pancreatic epithelium gives rise to all endocrine and exocrine cells of the mature organ. During organogenesis, the epithelial cells receive essential signals from the overlying mesenchyme. Previous studies, focusing on ex vivo tissue explants or complete knockout mice, have identified an important role for the mesenchyme in regulating the expansion of progenitor cells in the early pancreas epithelium. However, due to the lack of genetic tools directing expression specifically to the mesenchyme, the potential roles of this supporting tissue in vivo, especially in guiding later stages of pancreas organogenesis, have not been elucidated. We employed transgenic tools and fetal surgical techniques to ablate mesenchyme via Cre-mediated mesenchymal expression of Diphtheria Toxin (DT) at the onset of pancreas formation, and at later developmental stages via in utero injection of DT into transgenic mice expressing the Diphtheria Toxin receptor (DTR) in this tissue. Our results demonstrate that mesenchymal cells regulate pancreatic growth and branching at both early and late developmental stages by supporting proliferation of precursors and differentiated cells, respectively. Interestingly, while cell differentiation was not affected, the expansion of both the endocrine and exocrine compartments was equally impaired. To further elucidate signals required for mesenchymal cell function, we eliminated β-catenin signaling and determined that it is a critical pathway in regulating mesenchyme survival and growth. Our study presents the first in vivo evidence that the embryonic mesenchyme provides critical signals to the epithelium throughout pancreas organogenesis. The findings are novel and relevant as they indicate a critical role for the mesenchyme during late expansion of endocrine and exocrine compartments. In addition, our results provide a molecular mechanism for mesenchymal expansion and survival by identifying β-catenin signaling as an essential mediator of this process. These results have implications for developing strategies to expand pancreas progenitors and β-cells for clinical transplantation.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Gittes G. K. Developmental biology of the pancreas: a comprehensive review. Dev Biol. 2009;326:4–35. - PubMed
-
- Golosow N, Grobstein C. Epitheliomesenchymal interaction in pancreatic morphogenesis. Dev Biol. 1962;4:242–255. - PubMed
-
- Li Z, Manna P, Kobayashi H, Spilde T, Bhatia A, et al. Multifaceted pancreatic mesenchymal control of epithelial lineage selection. Dev Biol. 2004;269:252–263. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

